Touring multivariate data
SISBID 2025
https://github.com/dicook/SISBID
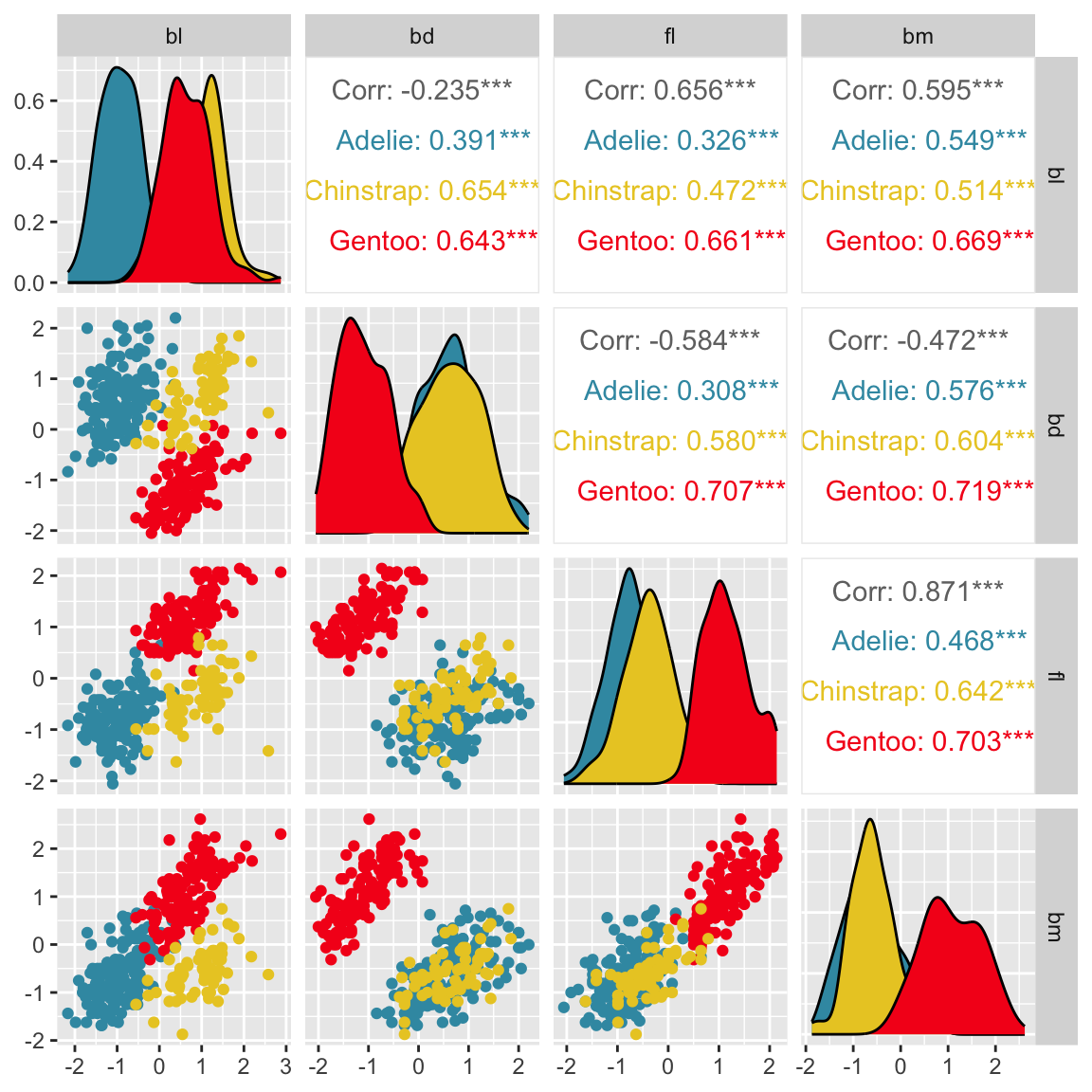
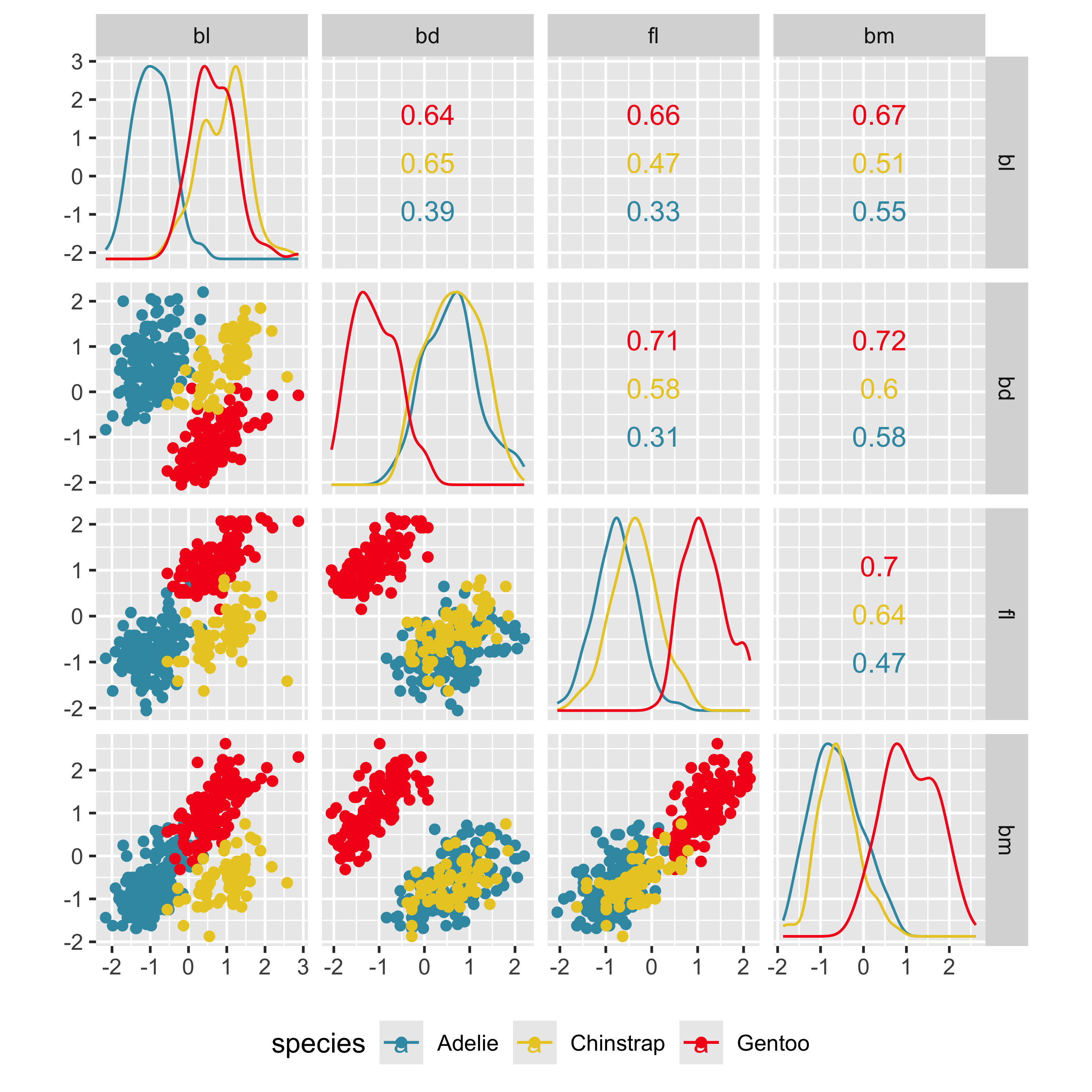
Pairwise plots
What don’t you see?
Unless you have tours, you’ll never know 🫣
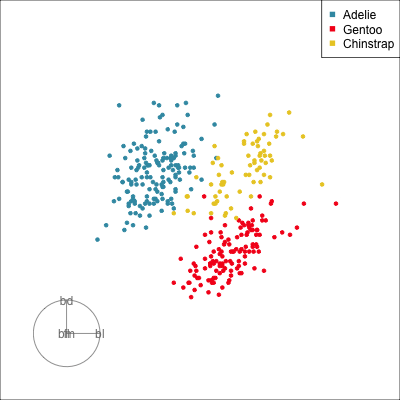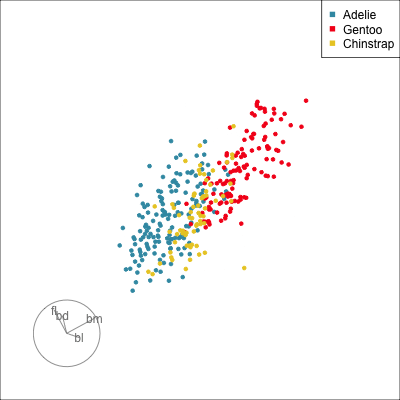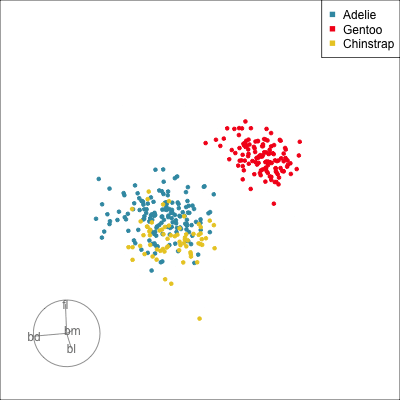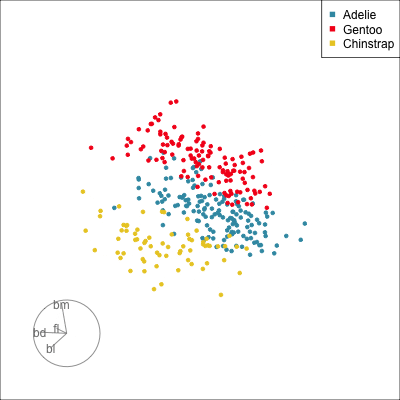
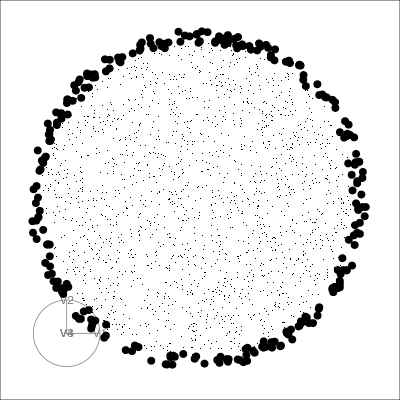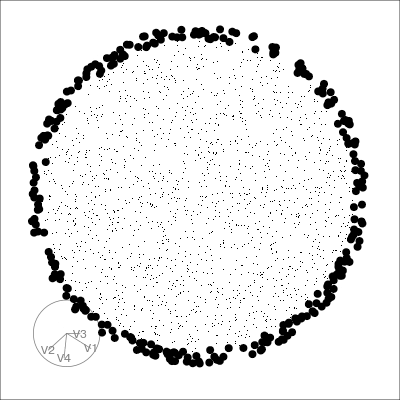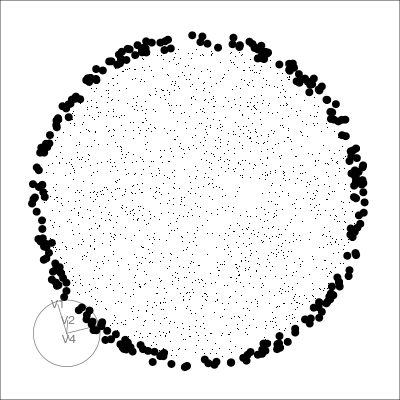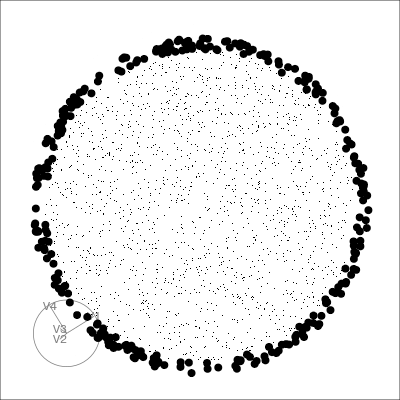
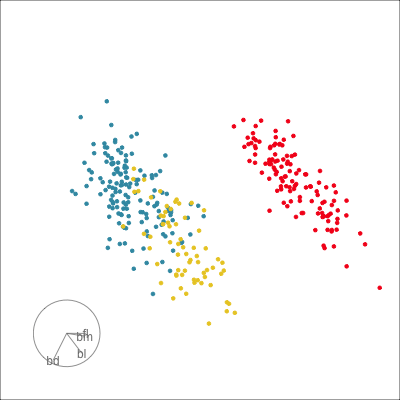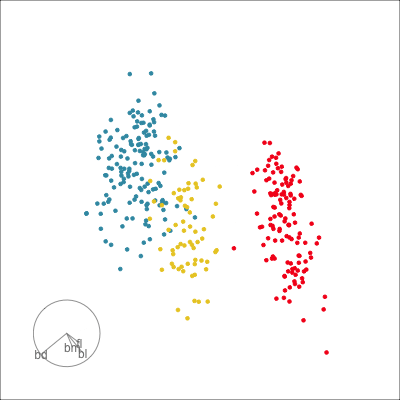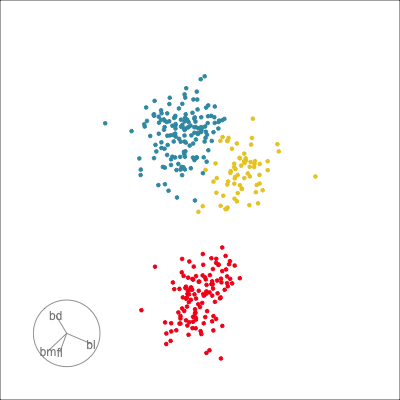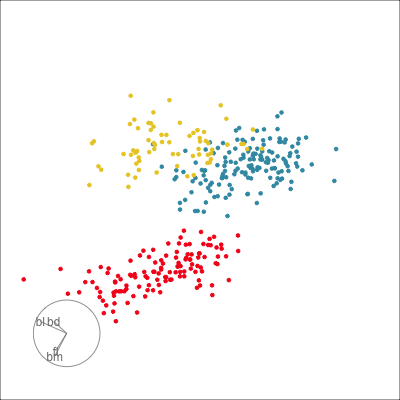
Our first tour
What patterns do you see?
What did you see?
- clusters ✅
- outliers ✅
- linear dependence ✅
- elliptical clusters with slightly different shapes ✅
- separated elliptical clusters with slightly different shapes ✅
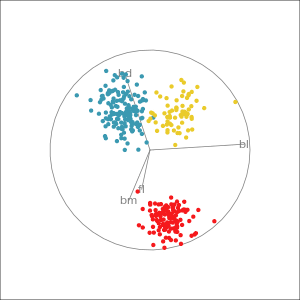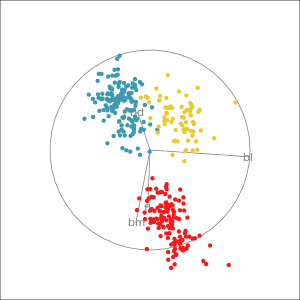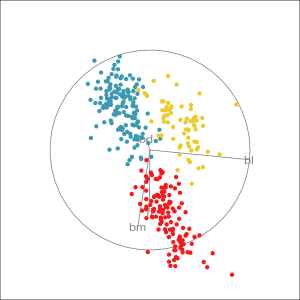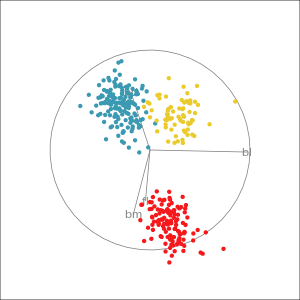
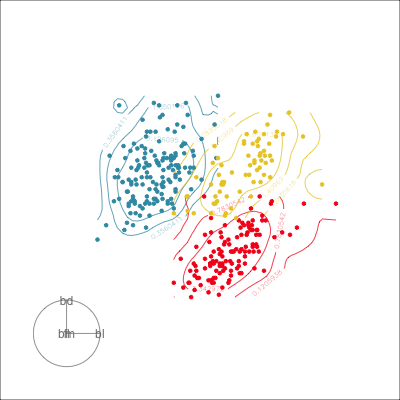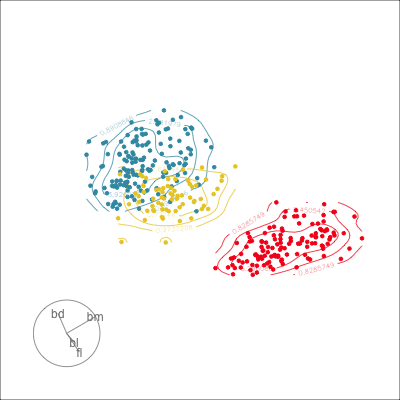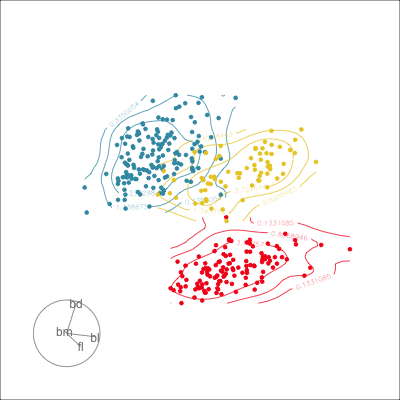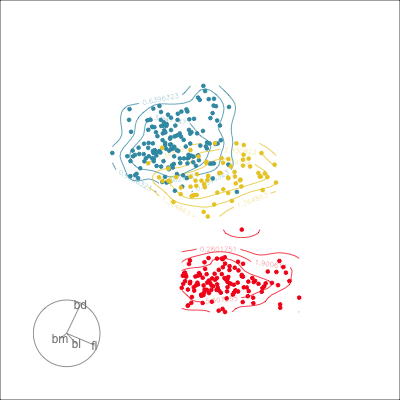
Which shows better separation?
What is a tour?
- a movie of low-dim projections
- constructed to come close to showing all possible low-dim projections
- a grand tour is a space-filling curve in the manifold of low-dim projections of high-dim data spaces.
\({\mathbf x}_i \in \mathcal{R}^p\), \(i^{th}\) data vector
\(F\) is a \(p\times d\) orthonormal basis
\(F'F=I_d\), where \(d\) is the projection dimension.
The projection of \({\mathbf x_i}\) onto \(F\) is \({\mathbf y}_i=F'{\mathbf x}_i\).
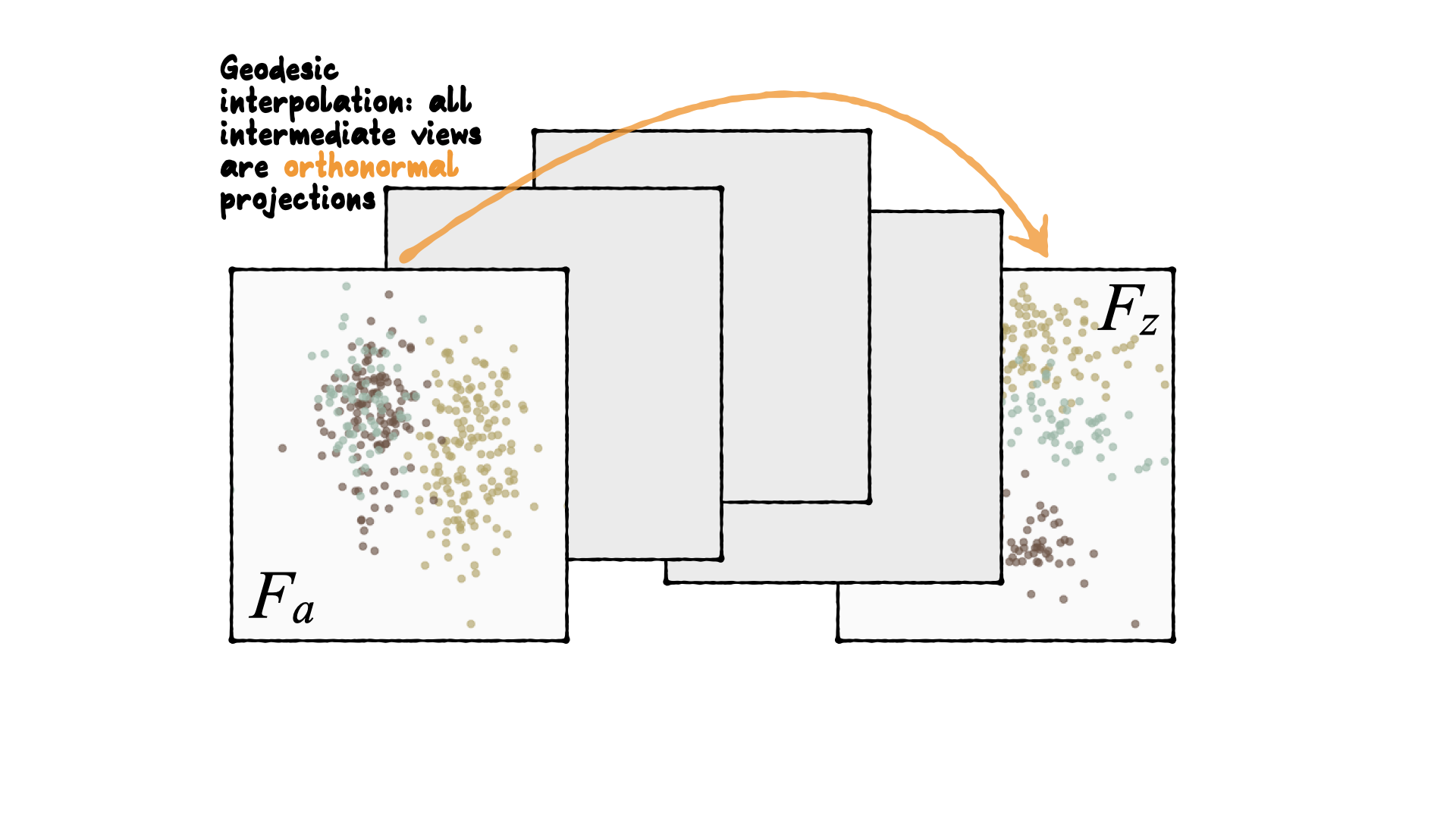
Tour is indexed by time, \(F(t)\), where \(t\in [a, z]\). Starting and target frame denoted as \(F_a = F(a), F_z=F(t)\).
The animation of the projected data is given by a path \({\mathbf y}_i(t)=F'(t){\mathbf x}_i\).
Geodesic interpolation b/w planes
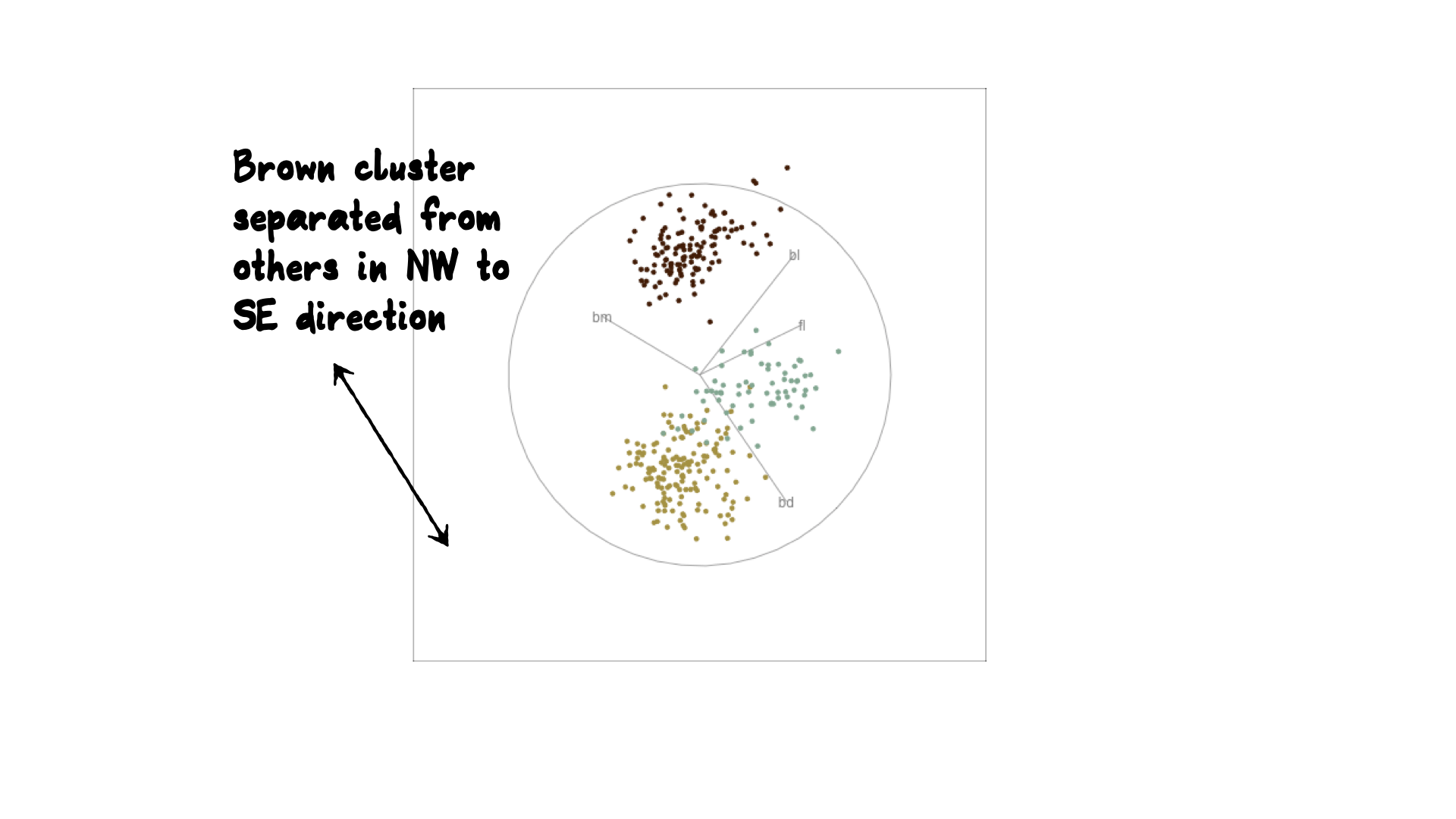
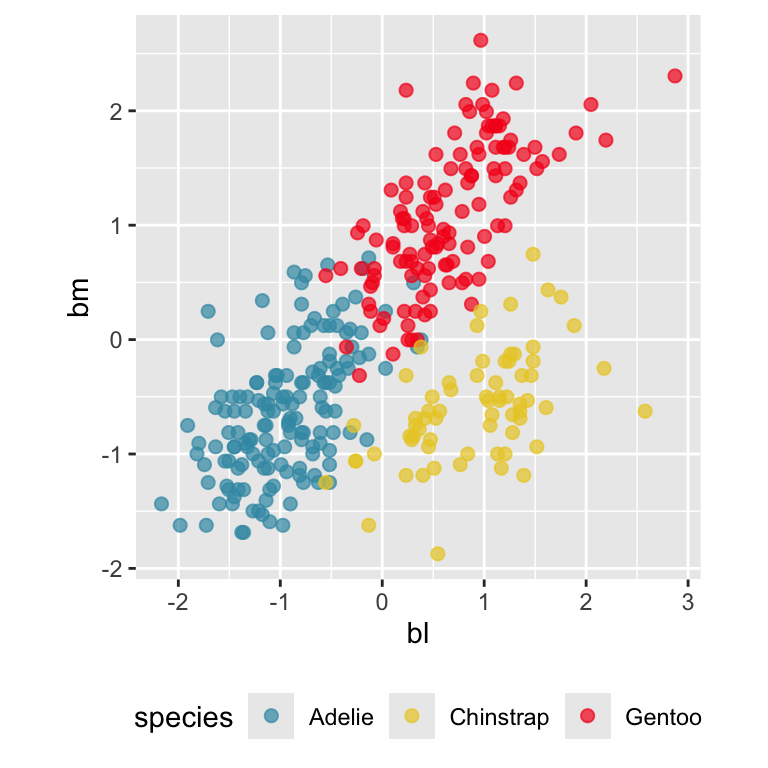
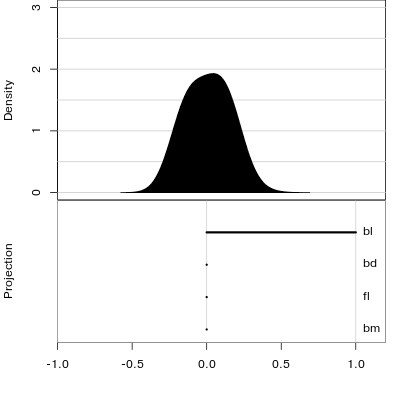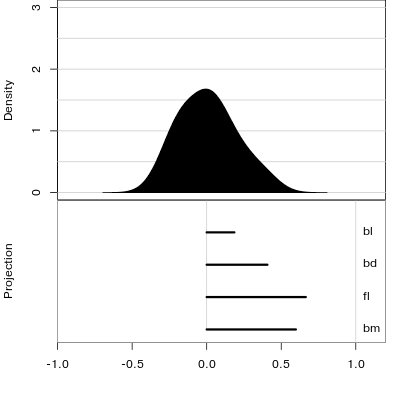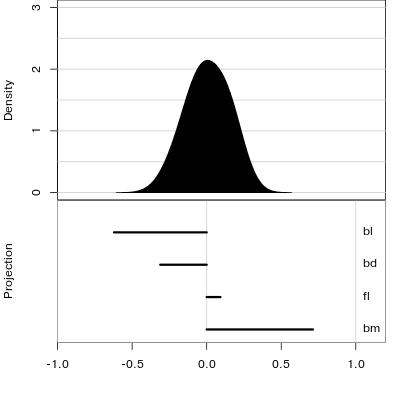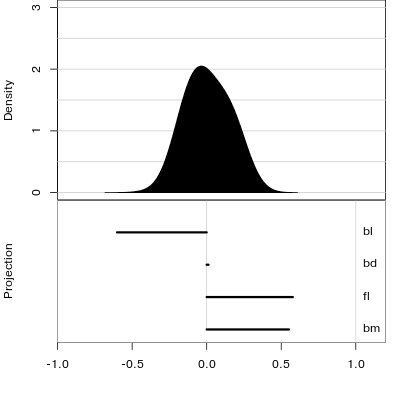
Reading axes - interpretation
Length and direction of axes relative to the pattern of interest
Reading axes - interpretation
Length and direction of axes relative to the pattern of interest
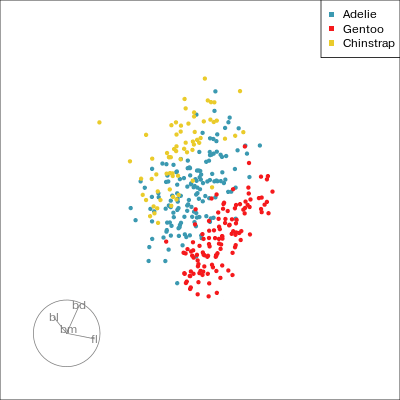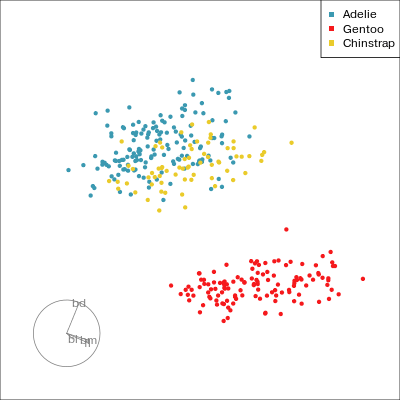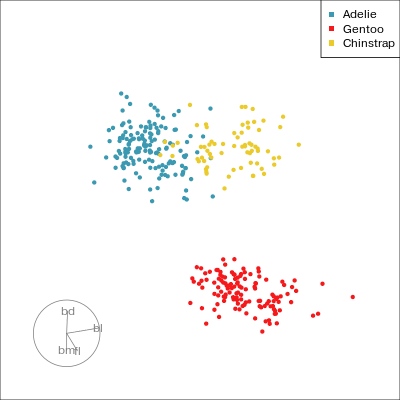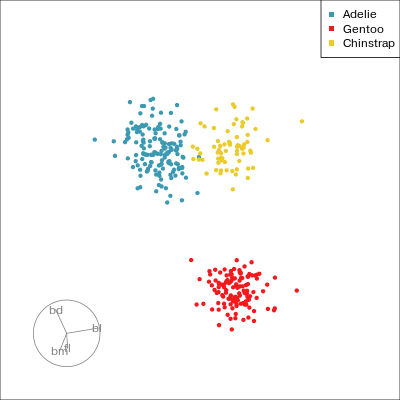
Understanding a tour
Understanding the projections
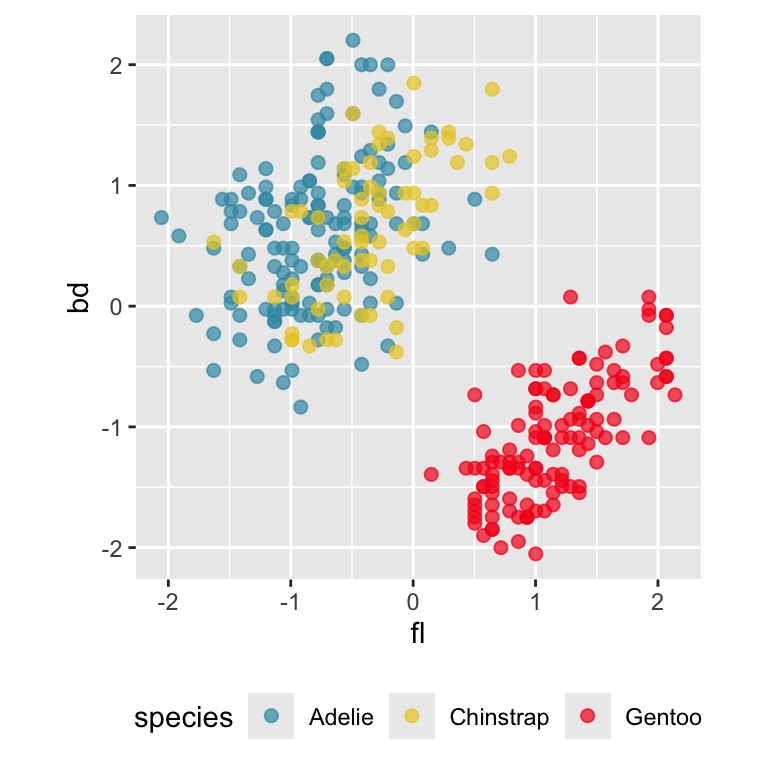
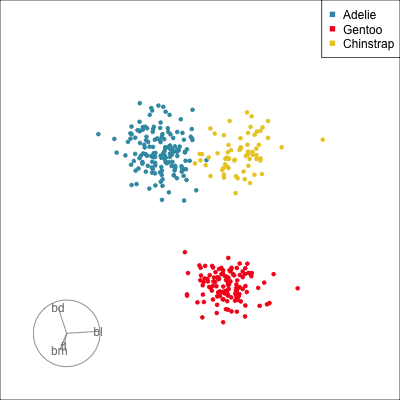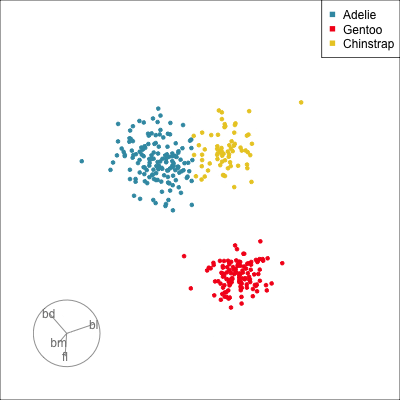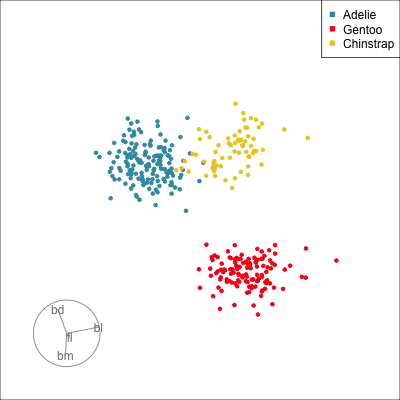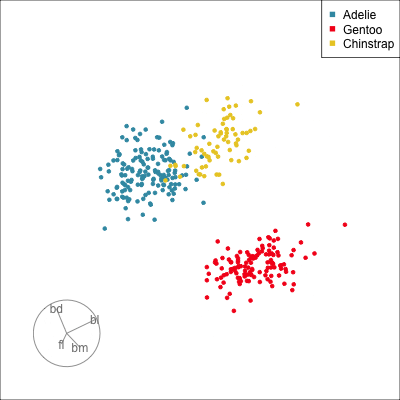
ggplot(penguins_std,
aes(x=fl, y=bd,
colour=species)) +
geom_point(alpha=0.7, size=2) +
scale_colour_discrete_divergingx(palette = "Zissou 1") +
theme(aspect.ratio=1,
legend.position="bottom") Gentoo from others in contrast of fl, bd
Difficulties in making interpretations
- There may be multiple and different combinations of variables that reveal similar structure. ☹️
- This is due to association between variables in the multivariate data.
- The tour can help to discover these, too. 😂
Other tour types
- guided: follows the optimisation path for a projection pursuit index.
- little: interpolates between all variables.
- local: rocks back and forth from a given projection, so shows all possible projections within a radius.
- dependence: two independent 1D tours
- frozen: fixes some variable coefficients, others vary freely.
- manual: control coefficient of one variable, to examine the sensitivity of structure this variable. (In the
spinifexpackage) - slice: use a section instead of a projection.
- sage: transform a 2D projection, to avoid data piling.
Guided tour
New target bases are chosen using a projection pursuit index function
\[\mathop{\text{maximize}}_{F}~g(xF) ~~~\text{ subject to } F \text{ being orthonormal}\]
holes: This is an inverse Gaussian filter, which is optimised when there is not much data in the center of the projection, i.e. a “hole” or donut shape in 2D.central mass: The opposite of holes, high density in the centre of the projection, and often “outliers” on the edges.LDA/PDA: An index based on the linear discriminant dimension reduction (and penalised), optimised by projections where the named classes are most separated.
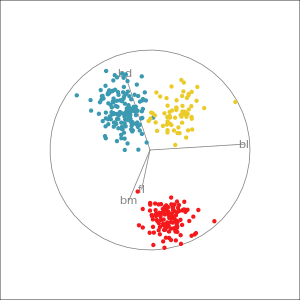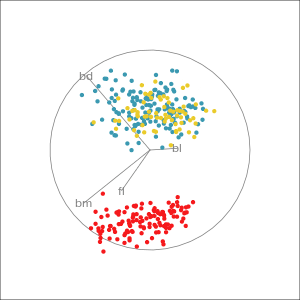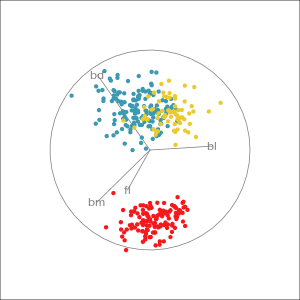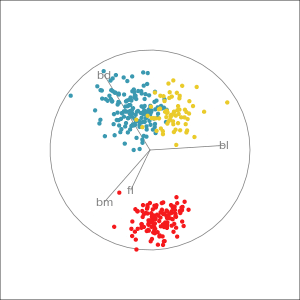
Manual tour
- start from best projection, given by projection pursuit
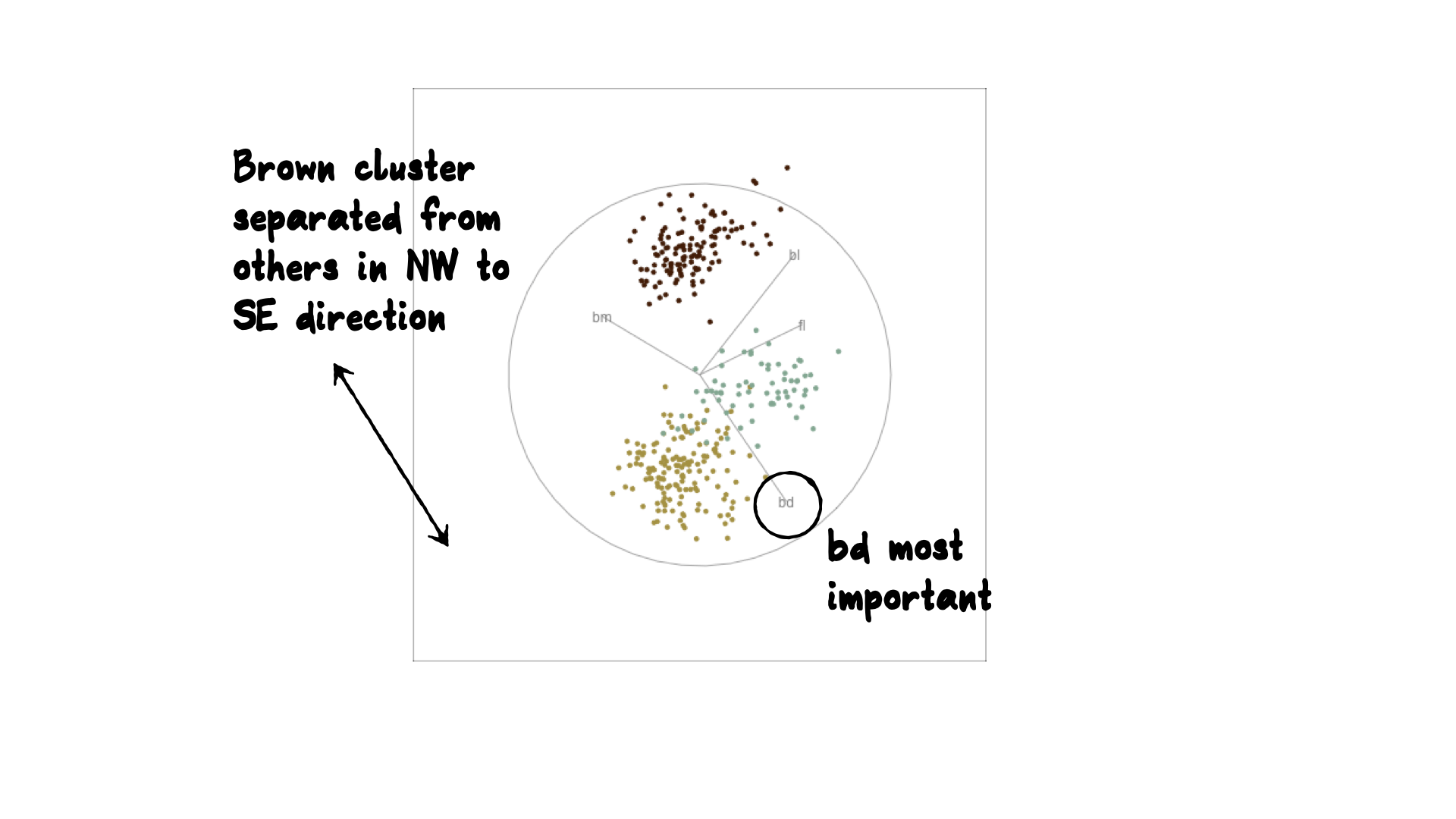
bdcontribution controlled- if
bdis removed from projection, Gentoo separation disappears bdis important for distinguishing Gentoo
# Check contribution of bl,
# change mvar to switch variables
animate_xy(penguins_std[,2:5],
radial_tour(as.matrix(best_proj), mvar = 2),
col = penguins_std$species)Manual tour
Local Tour
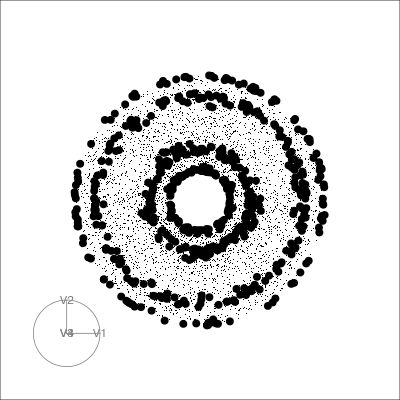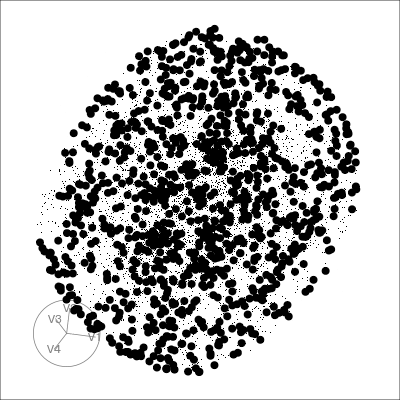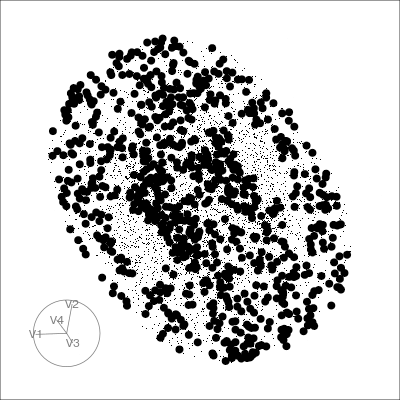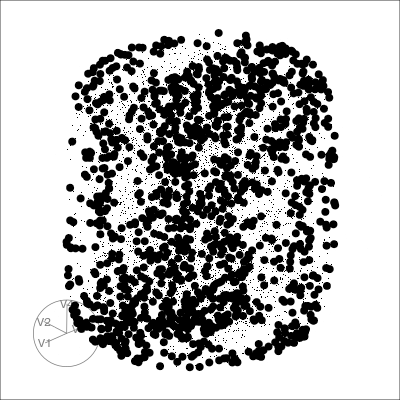
Geometric shapes with slice tour
Geometric shapes with slice tour
4D Torus
torus <- torus(p = 4, n = 5000, radius=c(8, 4, 1))$points %>% as_tibble()
animate_slice(torus, axes="bottomleft", half_range=0.8)PCA tour
Compute PCA, reduce dimension, show original variable axes in the reduced space.
penguins_pca <- prcomp(penguins_std[,2:5],
center = FALSE)
penguins_coefs <- penguins_pca$rotation[, 1:3]
penguins_scores <- penguins_pca$x[, 1:3]
animate_pca(penguins_scores, pc_coefs = penguins_coefs, col=penguins_std$species)render_gif(
penguins_scores, grand_tour(),
display_pca(pc_coefs = penguins_coefs,
col=penguins_std$species,
axes="bottomleft"),
"slides/images/penguins2d_pca.gif",
frames=100, width=400, height=400)Projection dimension and displays
Your turn
Using the sample code from the tour package, check how many clusters are in the example data.
library(tourr)
data(flea)
?animate_xy
# On a Mac, start quartz window with: quartz()
# On windows, start X11 window with: X11()
animate_xy(flea[, 1:6])
# RStudio graphics windows: may want to reduce frame rate
animate_xy(flea[, 1:6], fps=10)
# Also
animate_xy(flea[, -7], col = flea$species)
animate_xy(flea[, 1:6], tour_path = guided_tour(lda_pp(flea$species)), col=flea$species)Saving and sharing: Animated gif
Saving and sharing: single frame
Draw it with ggplot, and possibly pass to plotly.
load(here::here("data/p_tour_path.rda"))
penguins_pcti <- interpolate(
penguins_pct, 0.2)
f27 <- matrix(
penguins_pcti[,,27],
ncol=2)
p27 <- render_proj(
penguins_std[,2:5],
f27,
obs_labels=
penguins_std$species)Resources
- Cook and Laa (2025)
- Emerson et al (2013) The Generalized Pairs Plot, JCGS, 22:1, 79-91
- Natalia da Silva: PPForest and shiny app.
- Wickham et al (2011) tourr: An R Package for Exploring Multivariate Data with Projections, tourr R package
- Schloerke et al (2016) Escape from Boxland, the web site zoo and geozoo R package
- Spyrison and Cook (2020). spinifex: Manual Tours, Manual Control of Dynamic Projections of Numeric Multivariate Data.
- Stuart Lee liminal: tools to do linked brushing between tours and PCA/tSNE/PDS views
 This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.