ff_long <- french_fries |> pivot_longer(potato:painty, names_to = "type", values_to = "rating")
ff_lm <- lm(rating~type+treatment+time+subject,
data=ff_long)Wrangling Data and Models
SISBID 2025
https://github.com/dicook/SISBID
Outline
Models and model output
Functions such as lm, glm, lmer, glmmTMB … allow us to fit models
e.g. fit french fry rating with respect to all designed main effects:
summary, str, resid, fitted, coef, … allow us to extract different parts of a model for a linear model. Other model functions work differently … major headaches!
summary(ff_lm)
Call:
lm(formula = rating ~ type + treatment + time + subject, data = ff_long)
Residuals:
Min 1Q Median 3Q Max
-7.0729 -1.9674 -0.4644 1.5202 10.2192
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.19851 0.25792 4.647 3.50e-06 ***
typegrassy -1.15513 0.15572 -7.418 1.49e-13 ***
typepainty 0.70055 0.15577 4.497 7.11e-06 ***
typepotato 5.13322 0.15572 32.964 < 2e-16 ***
typerancid 2.03293 0.15572 13.055 < 2e-16 ***
treatment2 -0.05128 0.12051 -0.426 0.6705
treatment3 -0.04546 0.12056 -0.377 0.7062
time2 0.10194 0.21613 0.472 0.6372
time3 -0.01361 0.21613 -0.063 0.9498
time4 -0.10250 0.21613 -0.474 0.6353
time5 -0.13301 0.21690 -0.613 0.5398
time6 -0.04611 0.21613 -0.213 0.8311
time7 -0.24621 0.21628 -1.138 0.2550
time8 -0.11626 0.21658 -0.537 0.5914
time9 0.13188 0.22783 0.579 0.5627
time10 0.54635 0.22783 2.398 0.0165 *
subject10 1.71451 0.24389 7.030 2.48e-12 ***
subject15 -0.35911 0.24489 -1.466 0.1426
subject16 0.47519 0.24408 1.947 0.0516 .
subject19 2.01651 0.24389 8.268 < 2e-16 ***
subject31 1.49284 0.25098 5.948 2.98e-09 ***
subject51 1.87284 0.24389 7.679 2.07e-14 ***
subject52 0.19484 0.24389 0.799 0.4244
subject63 0.96051 0.24389 3.938 8.37e-05 ***
subject78 -0.58283 0.24389 -2.390 0.0169 *
subject79 -0.53702 0.25027 -2.146 0.0320 *
subject86 0.43543 0.25098 1.735 0.0828 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.9 on 3444 degrees of freedom
(9 observations deleted due to missingness)
Multiple R-squared: 0.3995, Adjusted R-squared: 0.395
F-statistic: 88.13 on 26 and 3444 DF, p-value: < 2.2e-16Tidying model output
The package broom (broom.mixed for glmmTMB) gets model results into a tidy format at different levels
- model level:
broom::glance(values such as AIC, BIC, model fit, …) - coefficients in the model:
broom::tidy(estimate, confidence interval, significance level, …) - observation level:
broom::augment(fitted values, residuals, predictions, influence, …)
Goodness of fit statistics: glance
glance(ff_lm)
# A tibble: 1 × 12
r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 0.400 0.395 2.90 88.1 0 26 -8607. 17270. 17442.
# ℹ 3 more variables: deviance <dbl>, df.residual <int>, nobs <int>Model estimates: tidy
ff_lm_tidy <- tidy(ff_lm)
head(ff_lm_tidy)
# A tibble: 6 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 1.20 0.258 4.65 3.50e- 6
2 typegrassy -1.16 0.156 -7.42 1.49e- 13
3 typepainty 0.701 0.156 4.50 7.11e- 6
4 typepotato 5.13 0.156 33.0 2.28e-207
5 typerancid 2.03 0.156 13.1 4.69e- 38
6 treatment2 -0.0513 0.121 -0.426 6.70e- 1Model diagnostics: augment
ff_lm_all <- augment(ff_lm)
glimpse(ff_lm_all)
Rows: 3,471
Columns: 12
$ .rownames <chr> "1", "2", "3", "4", "5", "6", "7", "8", "9", "10", "11", "1…
$ rating <dbl> 2.9, 0.0, 0.0, 0.0, 5.5, 14.0, 0.0, 0.0, 1.1, 0.0, 11.0, 6.…
$ type <chr> "potato", "buttery", "grassy", "rancid", "painty", "potato"…
$ treatment <fct> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ time <fct> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ subject <fct> 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 10, 10, 10, 10, 10, 10, 10, 1…
$ .fitted <dbl> 6.33172754, 1.19851228, 0.04338221, 3.23143977, 1.89905775,…
$ .resid <dbl> -3.43172754, -1.19851228, -0.04338221, -3.23143977, 3.60094…
$ .hat <dbl> 0.007910376, 0.007911506, 0.007910376, 0.007910376, 0.00790…
$ .sigma <dbl> 2.899486, 2.900008, 2.900081, 2.899554, 2.899426, 2.897111,…
$ .cooksd <dbl> 4.169299e-04, 5.086106e-05, 6.662863e-08, 3.696831e-04, 4.5…
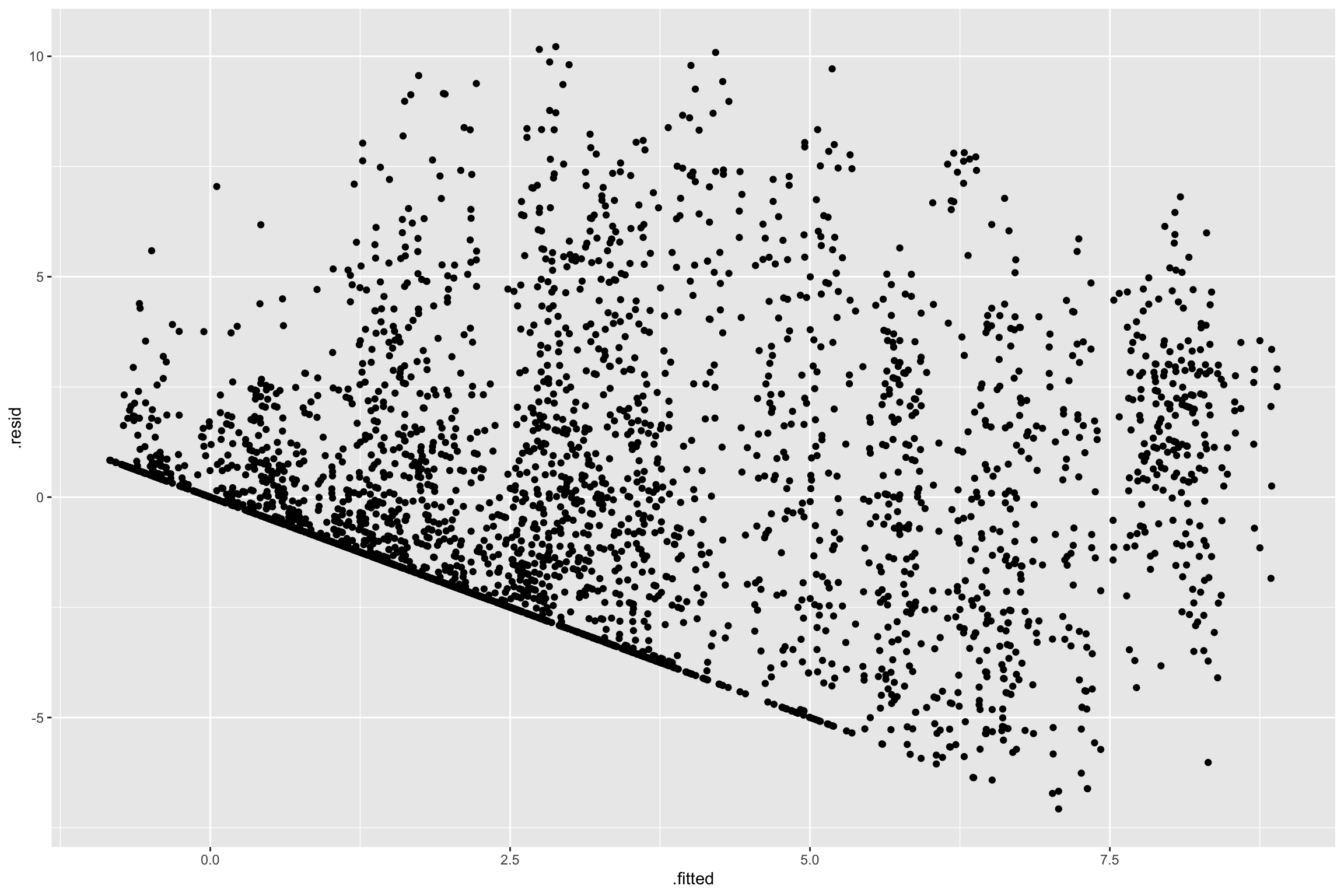
$ .std.resid <dbl> -1.18820206, -0.41497338, -0.01502067, -1.11885438, 1.24679…Residual plot
Adding random effects
Add random intercepts for each subject
fries_lmer <- lmer(rating~type+treatment+time + ( 1 | subject ),
data = ff_long)Your turn
- Run the code of the examples so far
The broom.mixed package provides similar functionality to mixed effects models as broom does for fixed effects models
- Try out the functionality of
broom.mixedon each level: model, parameters, and data - Augment the
ff_longdata with the model diagnostics - Plot the residuals from the random effects model
- Plot fitted values against observed values
dplyr verbs
There are five primary dplyr verbs, representing distinct data analysis tasks:
filter: Extract specified rows of a data framearrange: Reorder the rows of a data frameselect: Select specified columns of a data framemutate: Add new or transform a column of a data framesummarise: Create collapsed summaries of a data frame- (
group_by: Introduce structure to a data frame)
Filter
select a subset of the observations (horizontal selection)
load(here::here("data/french_fries.rda"))
french_fries |>
filter(subject == 3, time == 1) #<<
# A tibble: 6 × 9
time treatment subject rep potato buttery grassy rancid painty
* <fct> <fct> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 1 3 1 2.9 0 0 0 5.5
2 1 1 3 2 14 0 0 1.1 0
3 1 2 3 1 13.9 0 0 3.9 0
4 1 2 3 2 13.4 0.1 0 1.5 0
5 1 3 3 1 14.1 0 0 1.1 0
6 1 3 3 2 9.5 0 0.6 2.8 0 Arrange
order the observations (hierarchically)
french_fries |>
arrange(desc(rancid)) |> #<<
head()
# A tibble: 6 × 9
time treatment subject rep potato buttery grassy rancid painty
<fct> <fct> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 9 2 51 1 7.3 2.3 0 14.9 0.1
2 10 1 86 2 0.7 0 0 14.3 13.1
3 5 2 63 1 4.4 0 0 13.8 0.6
4 9 2 63 1 1.8 0 0 13.7 12.3
5 5 2 19 2 5.5 4.7 0 13.4 4.6
6 4 3 63 1 5.6 0 0 13.3 4.4Select
select a subset of the variables (vertical selection)
Summarise
summarize observations into a (set of) one-number statistic(s):
Summarise and group_by
french_fries |>
group_by(time, treatment) |>
summarise(mean_rancid = mean(rancid), sd_rancid = sd(rancid))
# A tibble: 30 × 4
# Groups: time [10]
time treatment mean_rancid sd_rancid
<fct> <fct> <dbl> <dbl>
1 1 1 2.76 3.21
2 1 2 1.72 2.71
3 1 3 2.6 3.20
4 2 1 3.9 4.37
5 2 2 2.14 3.12
6 2 3 2.50 3.38
7 3 1 4.65 3.93
8 3 2 2.90 3.77
9 3 3 3.6 3.59
10 4 1 2.08 2.39
# ℹ 20 more rowsLet’s use these tools
to answer these french fry experiment questions:
- Is the design complete?
- Are replicates like each other?
- How do the ratings on the different scales differ?
- Are raters giving different scores on average?
- Do ratings change over the weeks?
Completeness
If the data is complete it should be 12 x 10 x 3 x 2, that is, 6 records for each person in each week.
To check: tabulate number of records for each subject, time and treatment.
Your turn
How many values do we have for each subject? Check the help for function ?n
French Fries - completeness
n()
french_fries |>
group_by(subject) |>
summarize(n = n())
# A tibble: 12 × 2
subject n
<fct> <int>
1 3 54
2 10 60
3 15 60
4 16 60
5 19 60
6 31 54
7 51 60
8 52 60
9 63 60
10 78 60
11 79 54
12 86 54Other nice short cuts
instead of group_by(subject) |> summarize(n = n()) we can use:
group_by(subject) |> tally()count(subject)
Counts for subject by time
french_fries |>
na.omit() |>
count(subject, time) |>
pivot_wider(names_from="time", values_from="n")
# A tibble: 12 × 11
subject `1` `2` `3` `4` `5` `6` `7` `8` `9` `10`
<fct> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int>
1 3 6 6 6 6 6 6 6 6 6 NA
2 10 6 6 6 6 6 6 6 6 6 6
3 15 6 6 6 6 5 6 6 6 6 6
4 16 6 6 6 6 6 6 6 5 6 6
5 19 6 6 6 6 6 6 6 6 6 6
6 31 6 6 6 6 6 6 6 6 NA 6
7 51 6 6 6 6 6 6 6 6 6 6
8 52 6 6 6 6 6 6 6 6 6 6
9 63 6 6 6 6 6 6 6 6 6 6
10 78 6 6 6 6 6 6 6 6 6 6
11 79 6 6 6 6 6 6 5 4 6 NA
12 86 6 6 6 6 6 6 6 6 NA 6How do scores change over time?
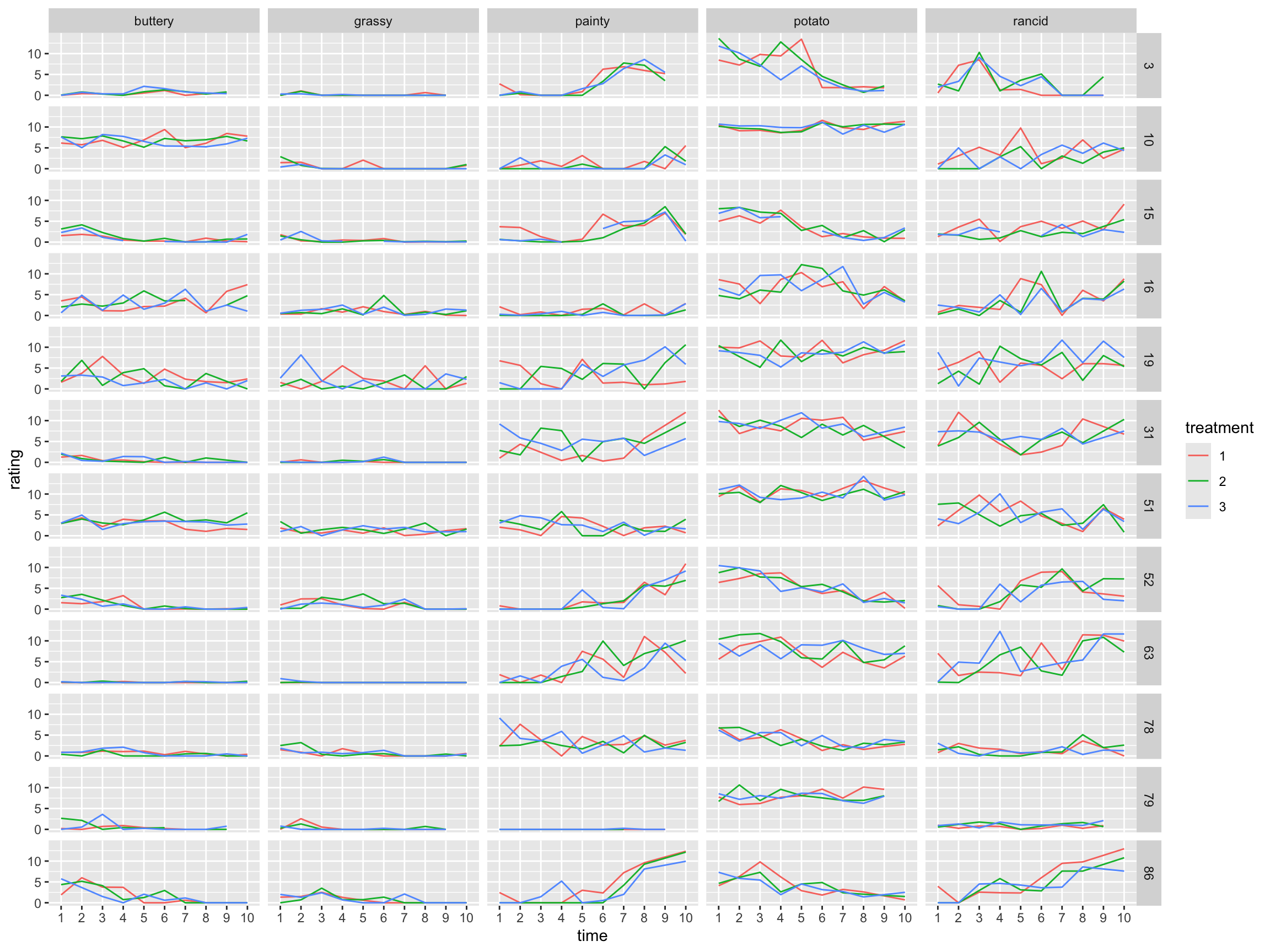
Your Turn
Get summary of ratings over replicates and connect the dots for a picture as below:
References
- Posit cheatsheets
- Wickham (2007) Reshaping data
- broom vignettes, David Robinson
- Wickham (2011) Split-Apply-Combine
 This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.