install.packages("nullabor")Preamble
SISBID 2025
https://github.com/dicook/SISBID
Interactive Sessions
We will be using breakout rooms throughout the class
- 4-6 people in each room
- at the end of the time you will be ‘zoomed back’ into the main session
- … if you get disconnected, just log back into the Zoom session and we will get you back to your group.
Let’s try this!
Your turn: meet the class! 🔮 👽 👼
We are going to break into three groups, you’ll be with either Heike, Susan and Di.
Find out:
- What is your favorite data plot?
- What are you hoping to learn from this workshop?
- What type of data are you often working with?
- Is there some analysis task that you have found really hard?
Datavis module logistics
- Website: https://dicook.github.io/SISBID/
- Zoom
- Sessions will be recorded and links will be made available via Slack
- Please remember to mute when not speaking
- In case of bandwidth issues, try turning off your camera
- Questions
- Zoom chat and Slack are both good places to ask questions
- R cloud link available from website & slack in case of technical problems
Your turn: Course materials
All materials at https://github.com/dicook/SISBID
- Download
SISBID_day1.zip(a zip archive). This contains all- the code files,
qmdfiles,- and data for day 1 of this workshop.
- Unzip the file Windows user: use extract, don’t just double-click
- Double-click
SISBID.Rproj. The R project helps organise your work over these next few days.
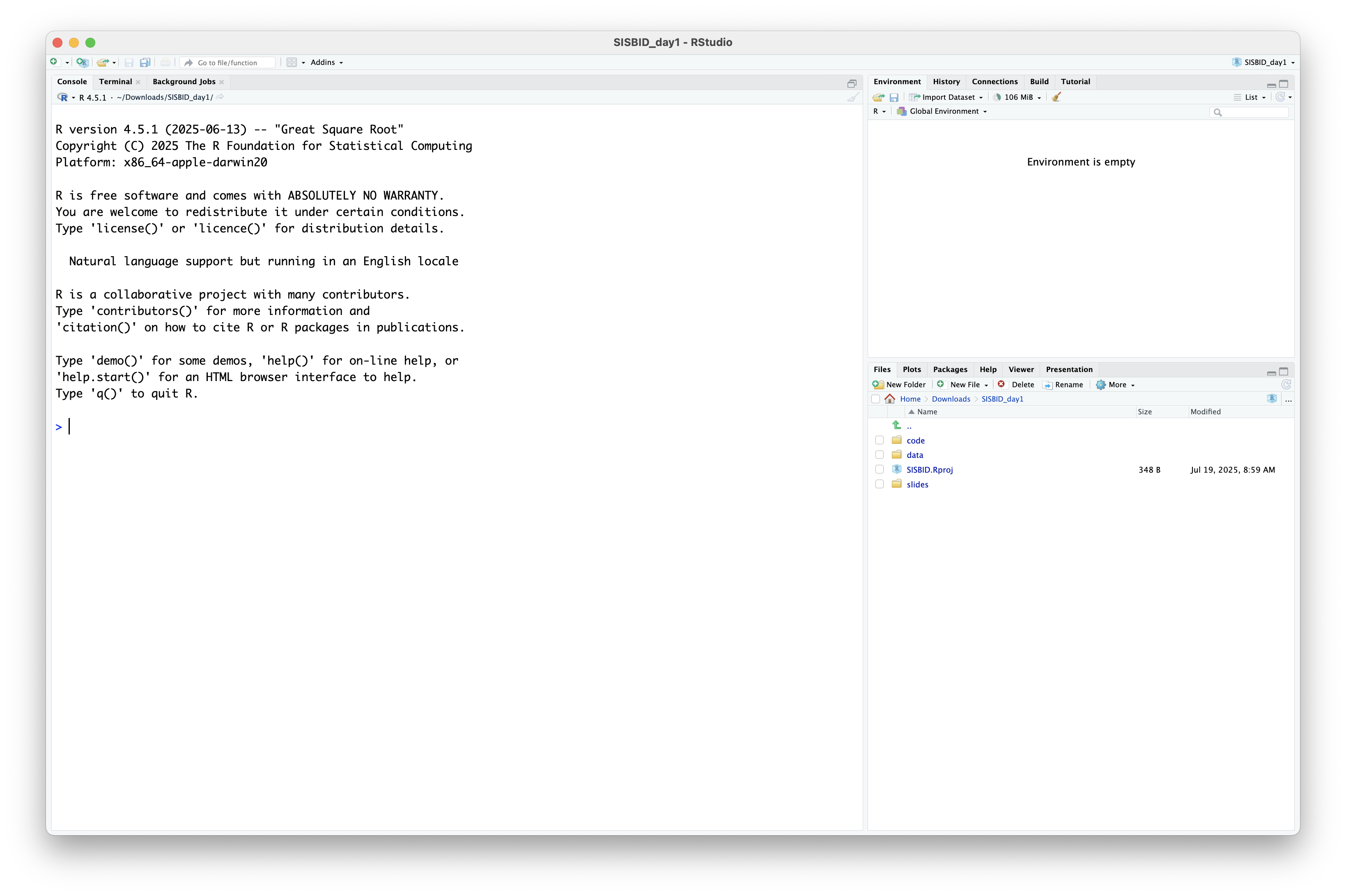
What’s in the zip?
.Rfile of code, one for each module of the course, use this to follow alongdatafolder containing data sets used in the courseSISBID.Rprojfor starting RStudio in your course working directory
Please run, edit and change the code as we go! Experiment, break and fix.
Check your R setup
“If R were an airplane, RStudio would be the airport”
Julie Lowndes, Openscapes
R package locations:
CRAN
Bioconductor
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("bigPint")Github
remotes::install_github("wmurphyrd/fiftystater")Code to install all packages required for the course is available on course website.
Resources
- RStudio cheatsheets
- Q/A site: http://stackoverflow.com
 This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.