| time | topic |
|---|---|
| 1:30-1:45 | Why, philosophy and benefits |
| 1:45-2:05 | Organising data to map variables to plots |
| 2:05-2:35 | Making a variety of plots |
| 2:35-3:00 | Do but don’t, and cognitive principles |
| 3:00-3:30 | BREAK |
Creating data plots for effective decision-making using statistical inference with R
Monash University
Session 1: Making effective plots using ggplot2’s grammar of graphics
Outline
Why (1/6)
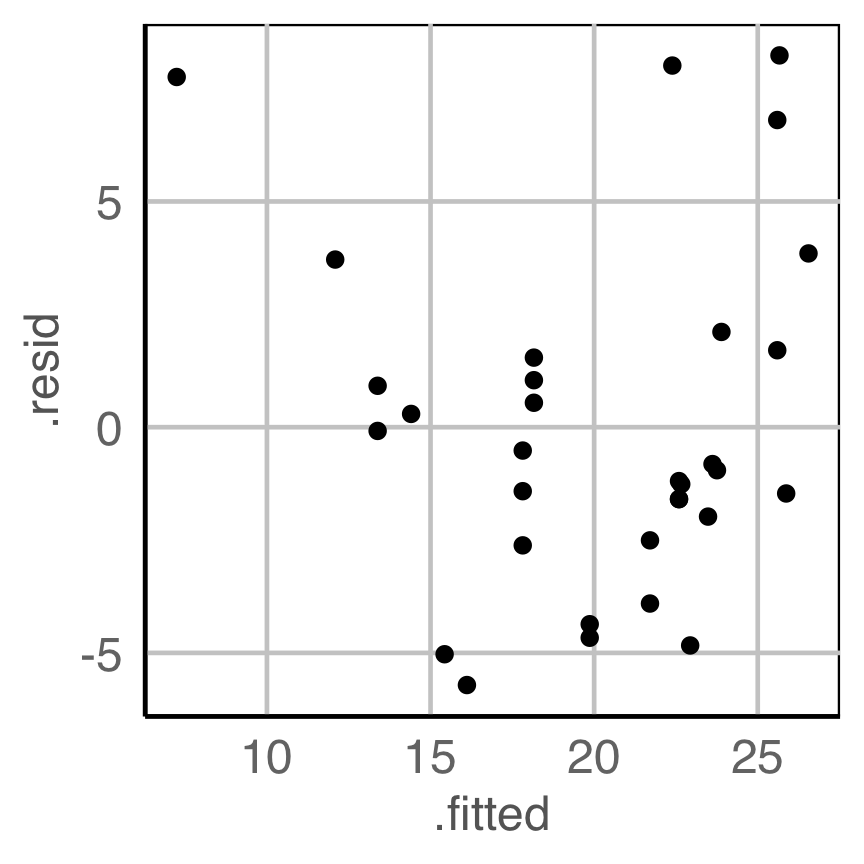
Is there any pattern in the residuals that indicate a problem with the model fit?
Do we need to change the model specification?
Why (2/6)
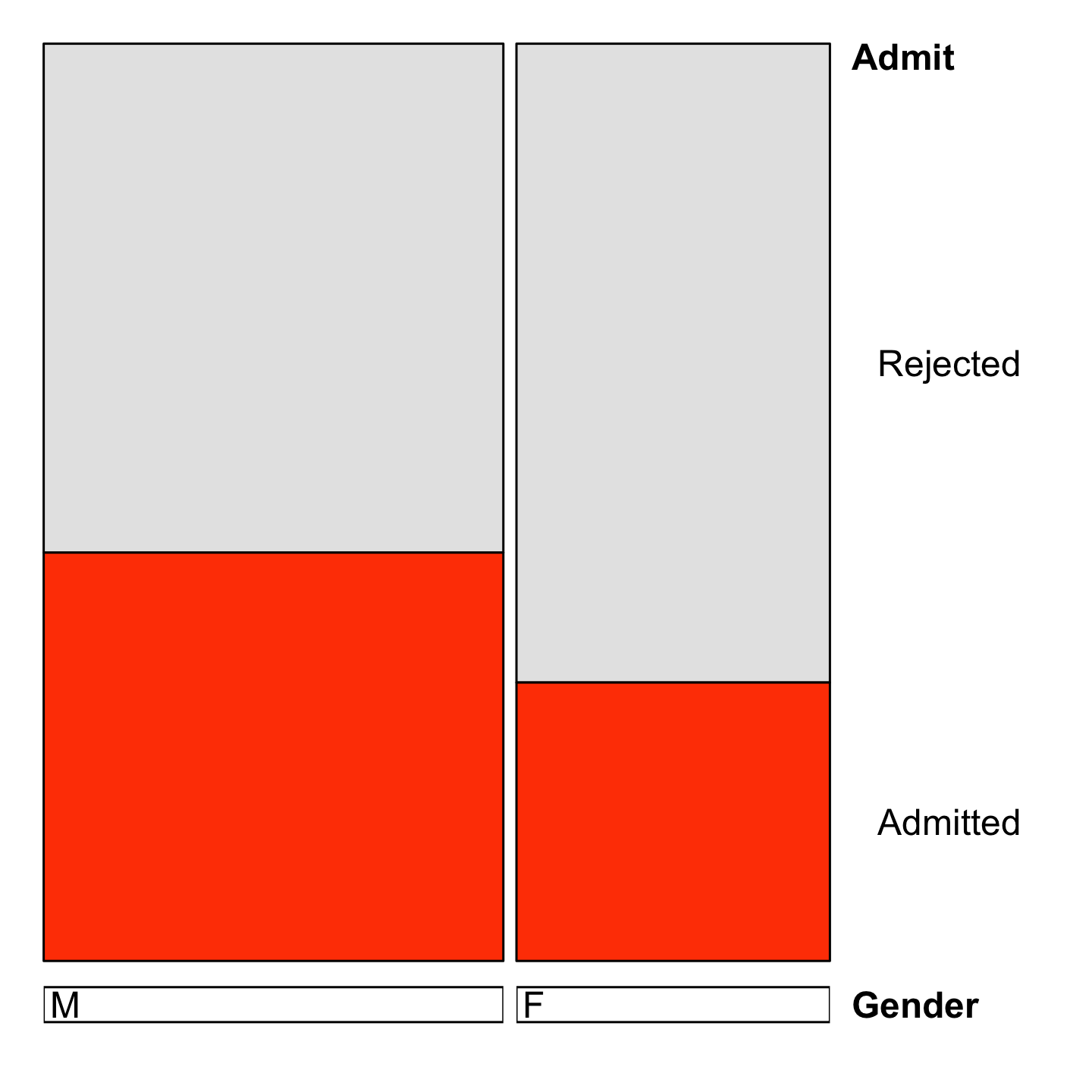
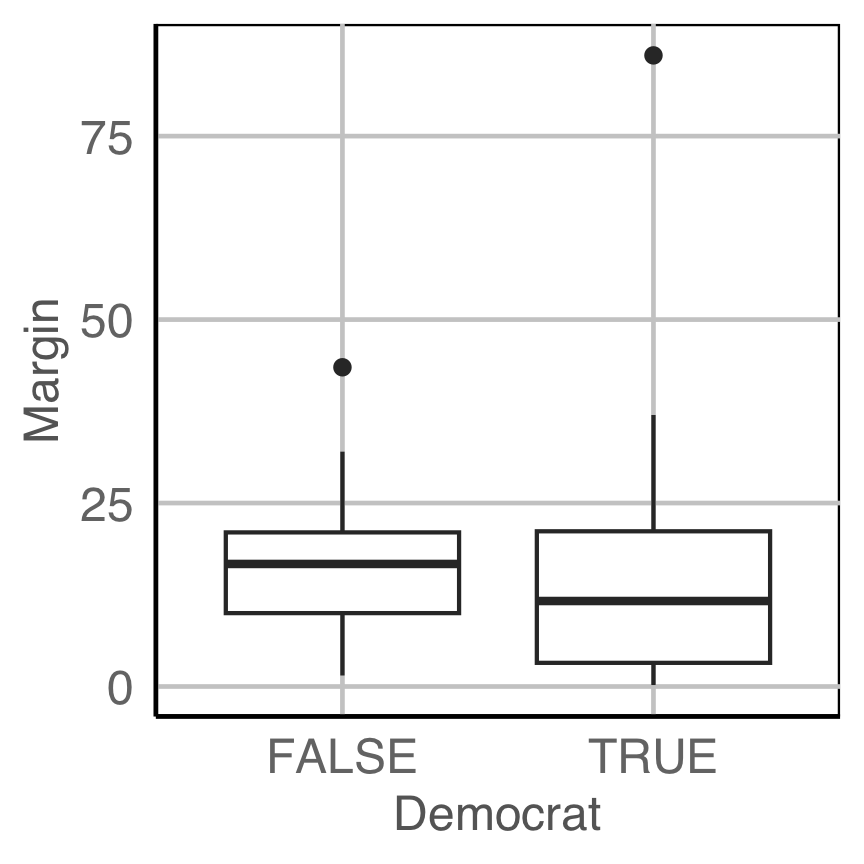
Is there a bias in admissions?
Why (3/6)
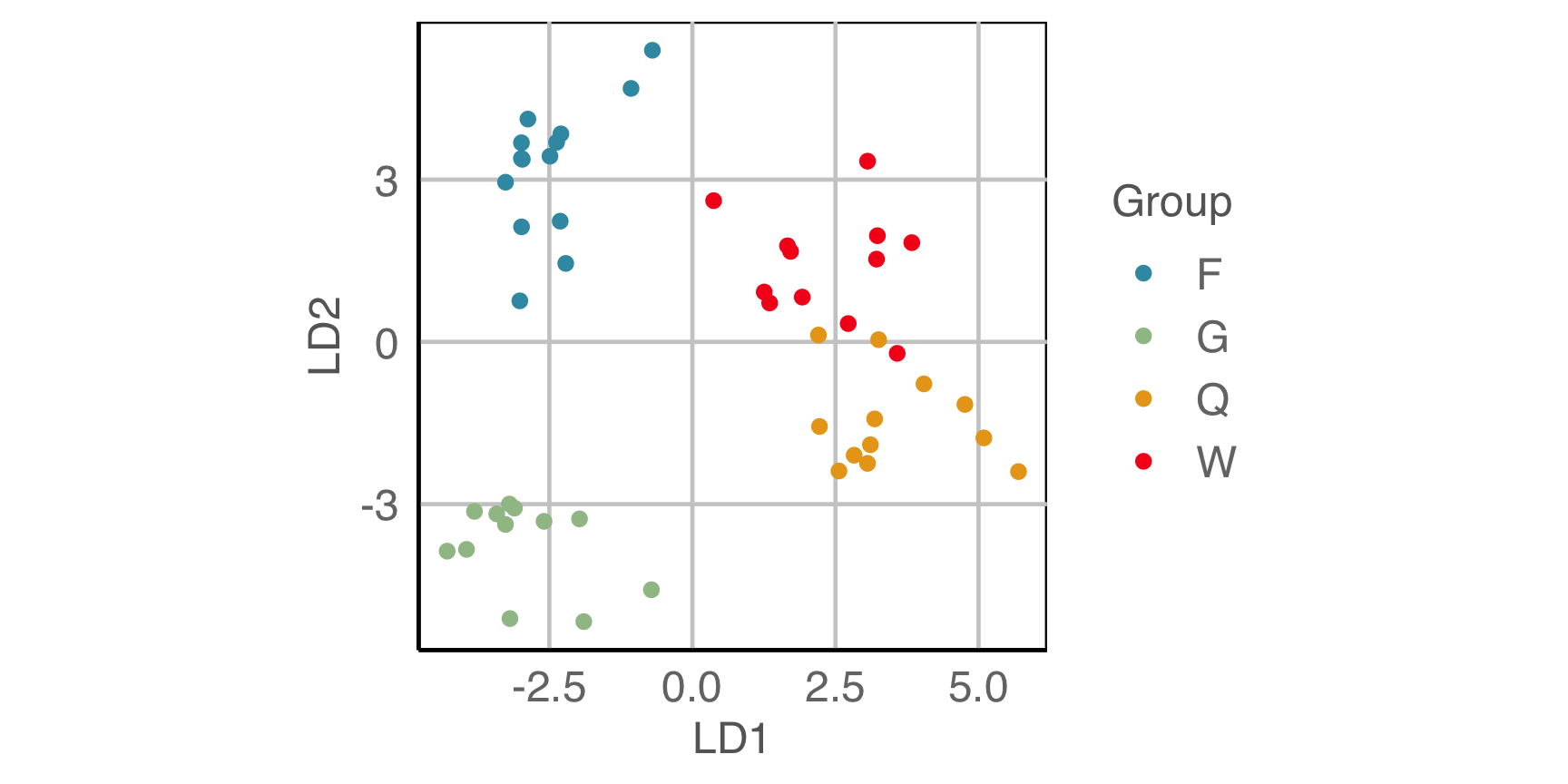
Is there a difference between the species?
Why (4/6)
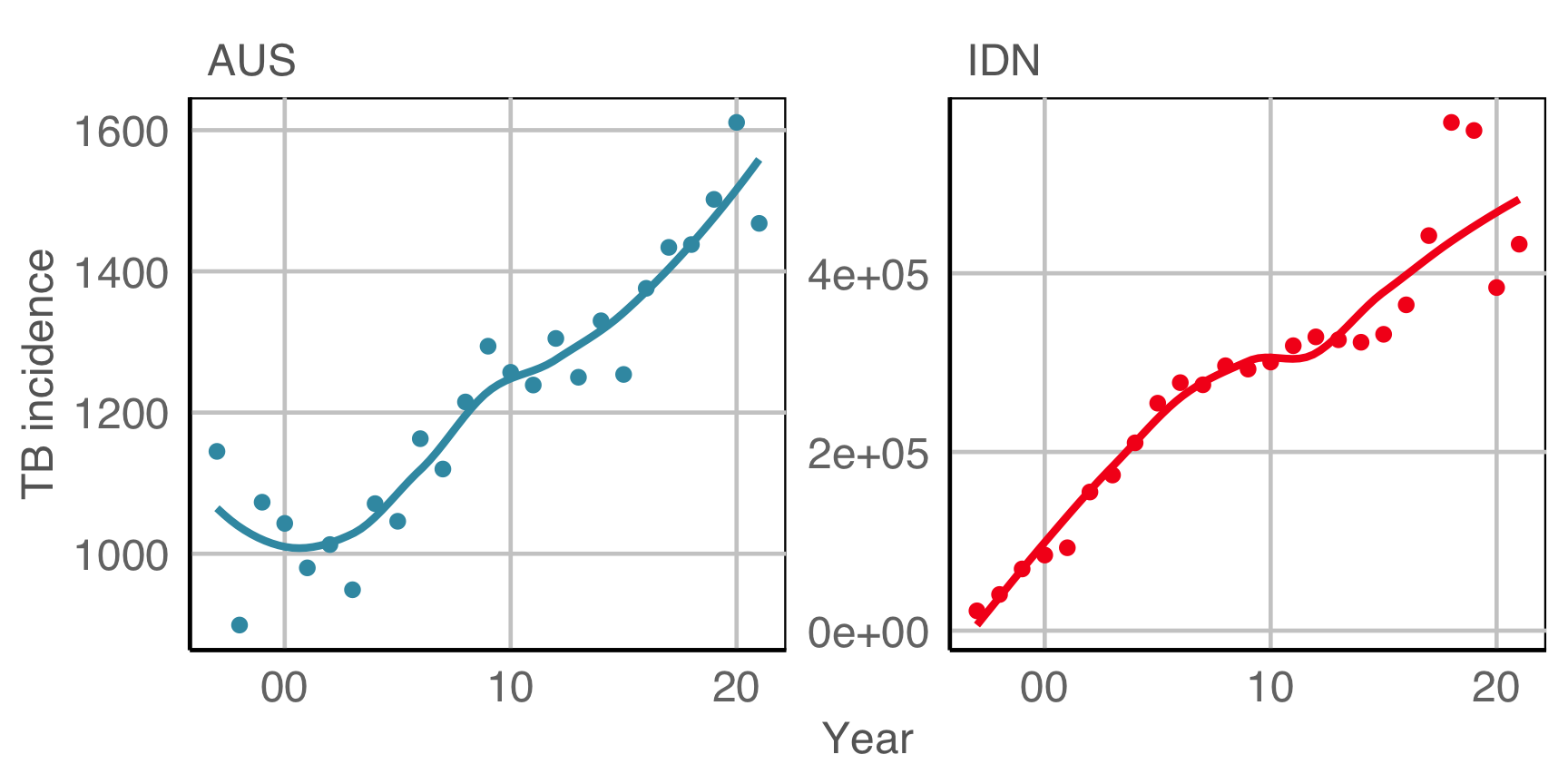
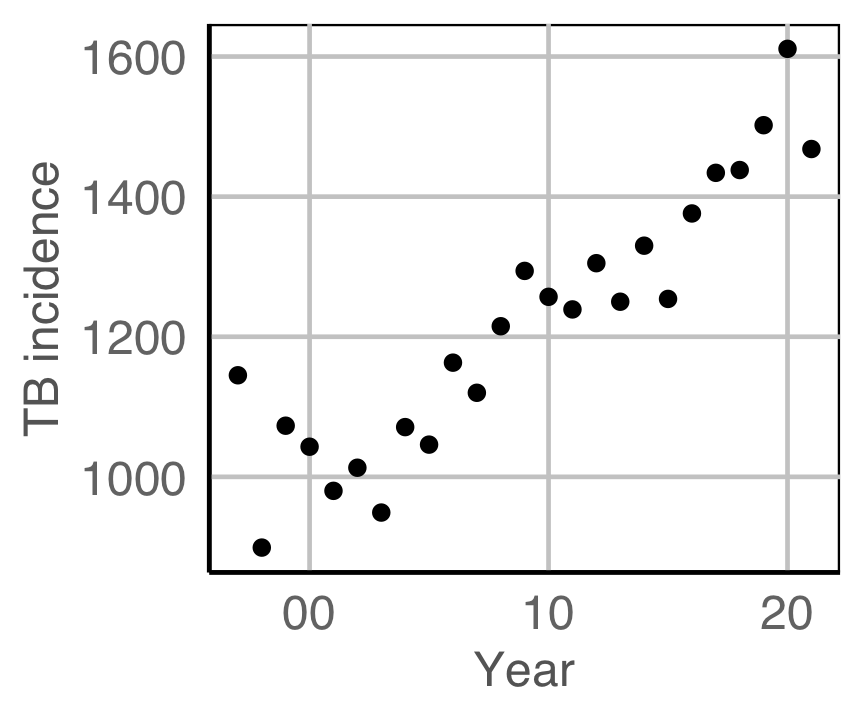
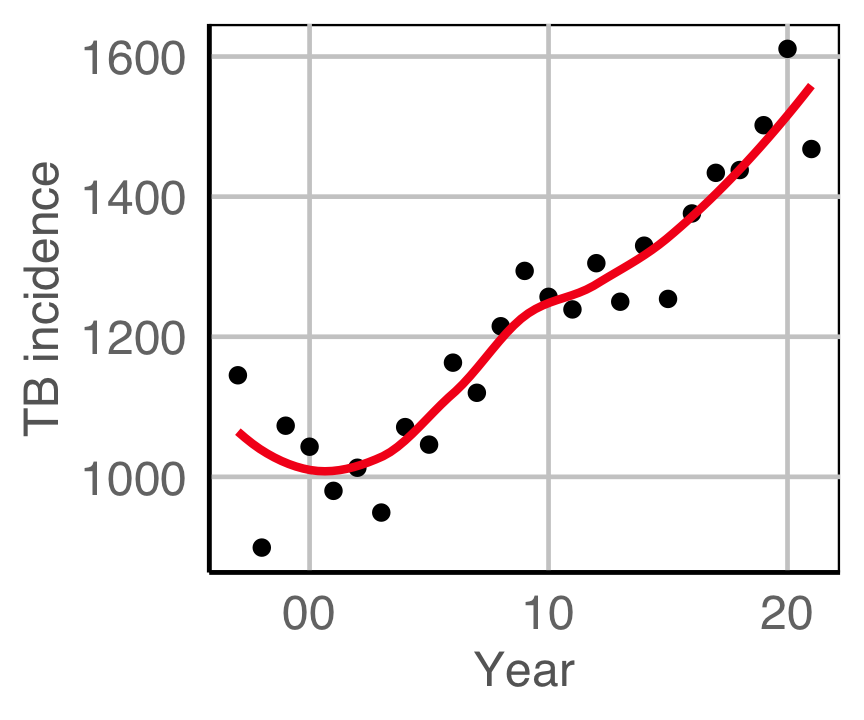
Is TB getting worse? (In Australia and Indonesia)
(From the World Health Organisation (WHO)]
Why (5/6)
Which is the best display to answer the previous question?
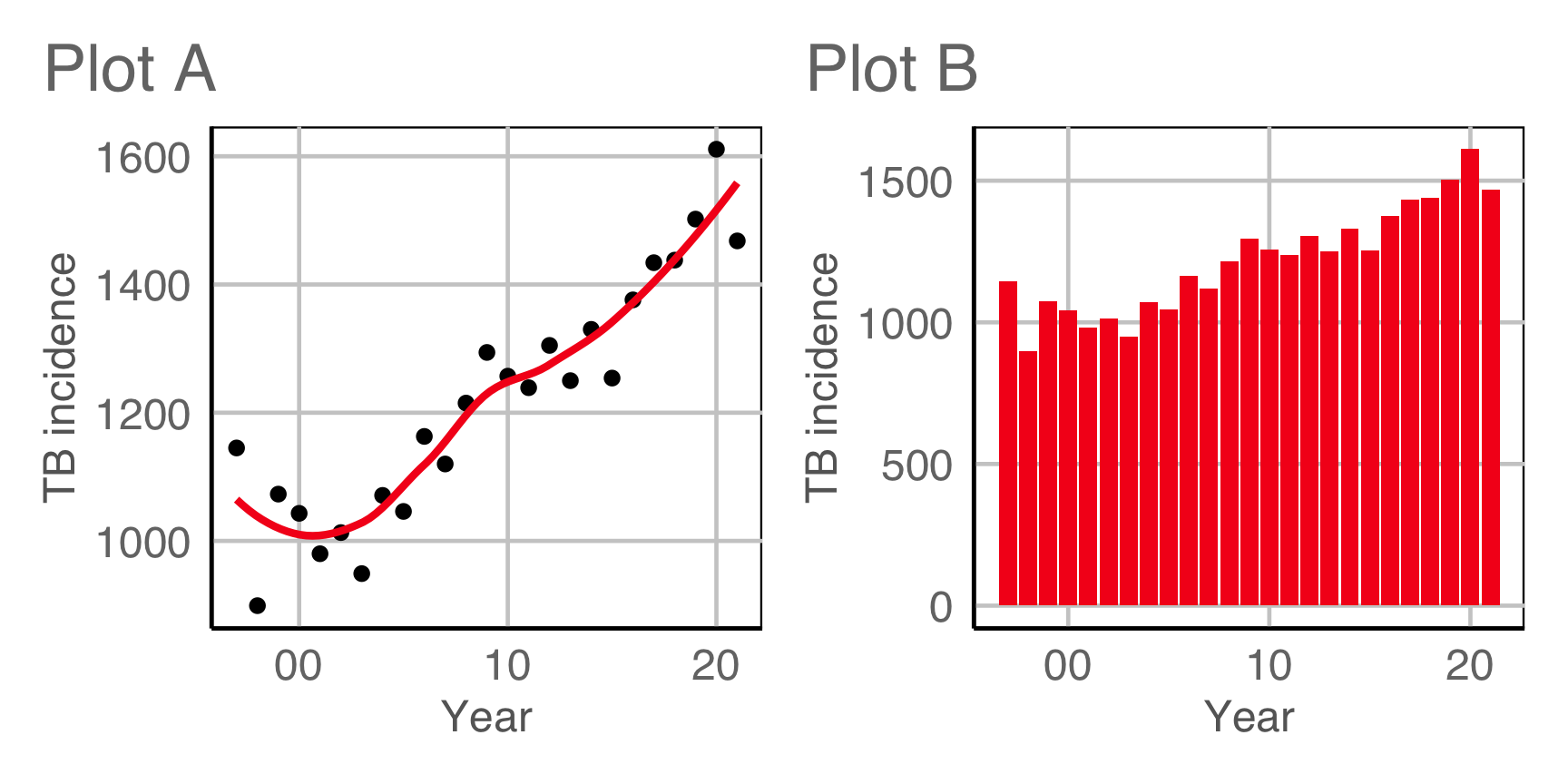
Why (6/6)
Which is the best display to answer: what is distribution of thyroid cancer across Australia?
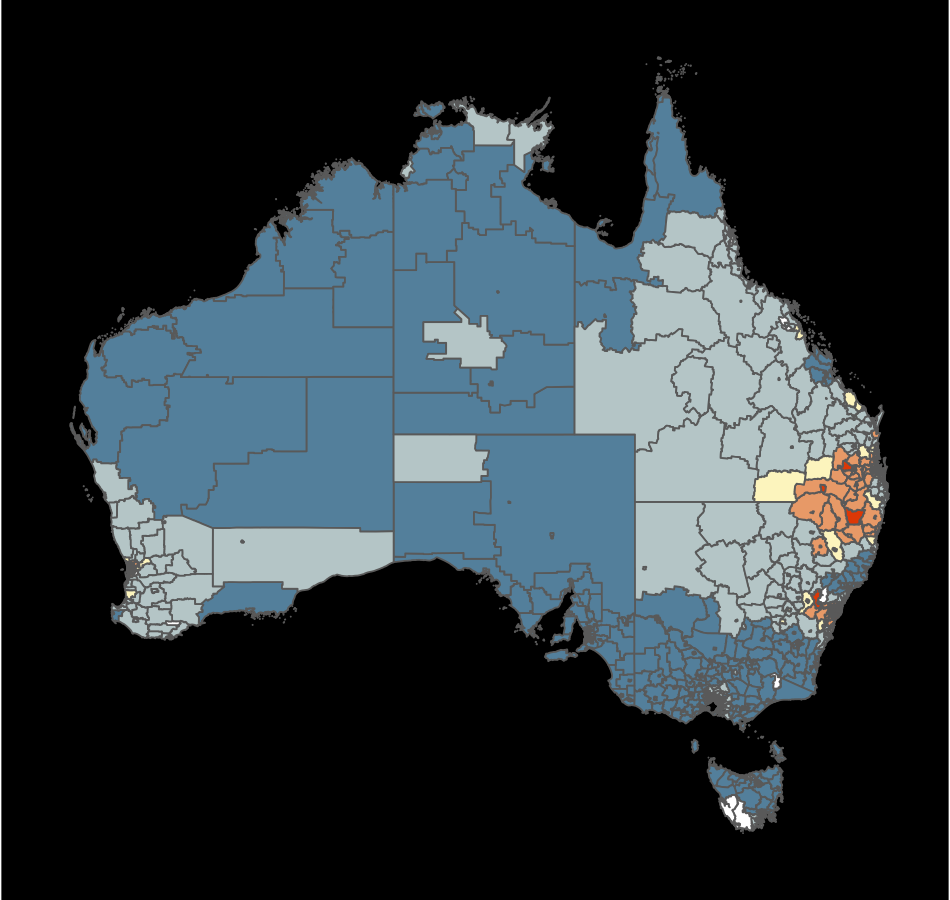
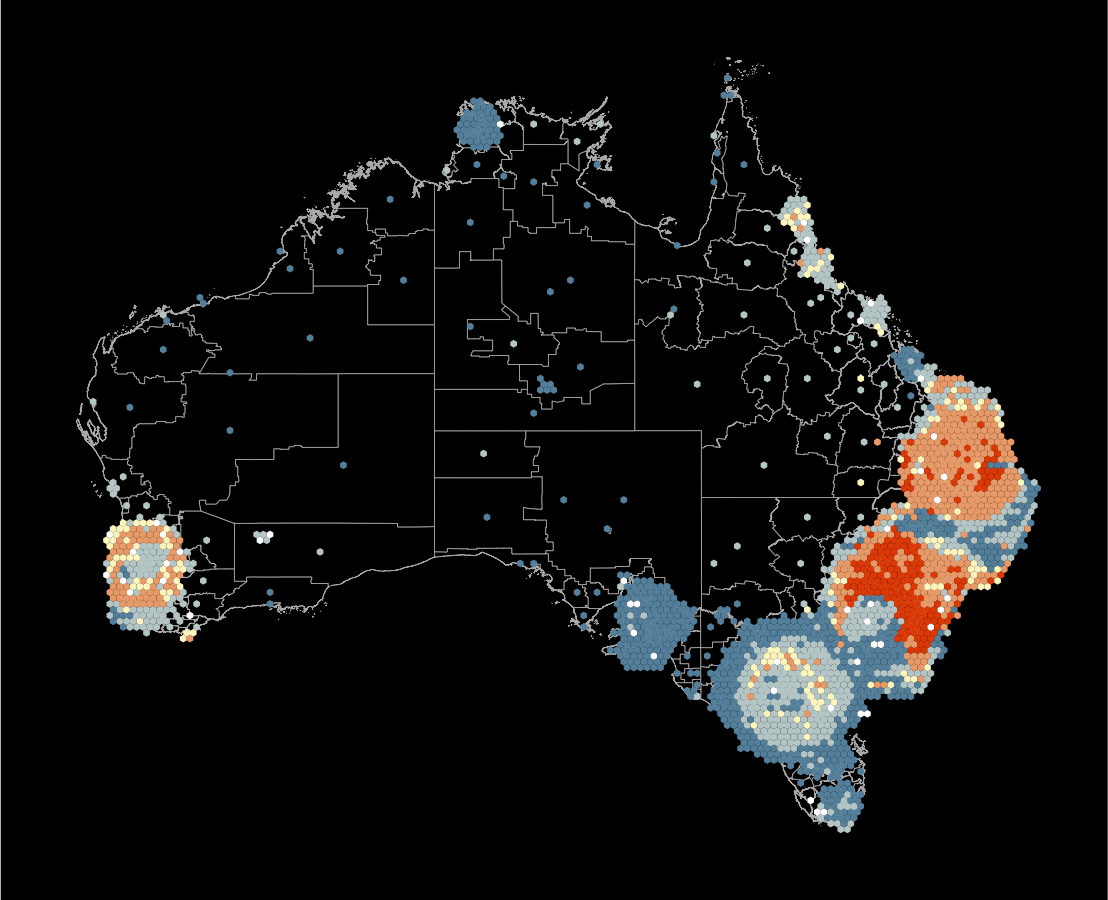
What’s the goal?
Reading data plots is subjective.
Making decisions based on data visualisations is common, where we need to be objective .
It is possible, and here is how we do that …
These are the tools you need
space
Organising your data to enable mapping variables to graphical elements
Tidy data
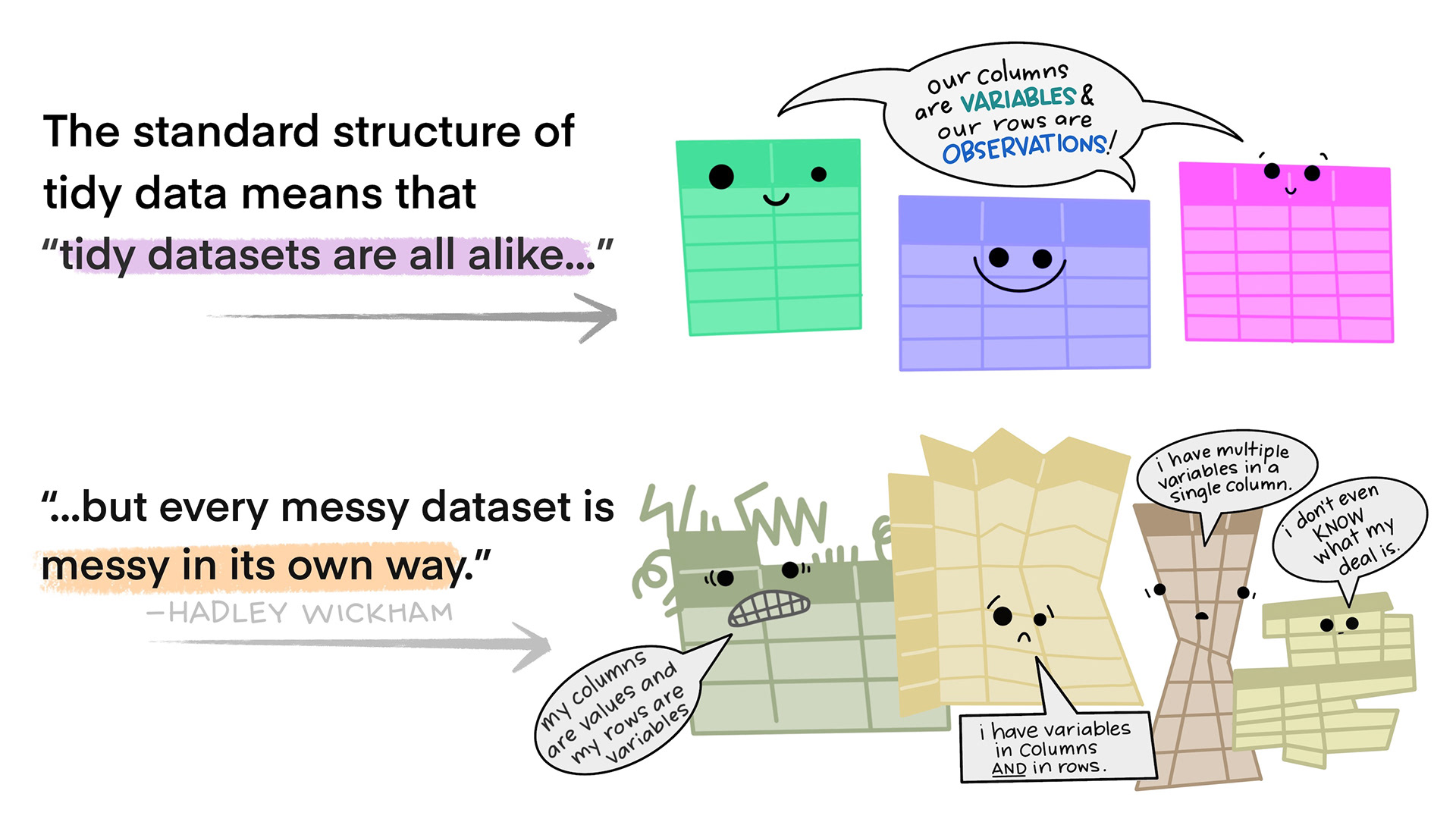
- Each variable forms a column
- Each observation forms a row
- Each type of observational unit forms a table. If you have data on multiple levels (e.g. data about houses and data about the rooms within those houses), these should be in separate tables.
Illustrations from the Openscapes blog Tidy Data for reproducibility, efficiency, and collaboration by Julia Lowndes and Allison Horst
YOUR TURN
For each of the following data discuss whether it is in tidy form.
Data 1: tuberculosis from WHO
Data 2: Graduate programs
Data 3: GHCN weather station records
Answers
Data 1:
Not in tidy form
- iso3
- year
- gender
- age
Data 2:
It’s in tidy form!
- subject
- inst
- AvNumPubs
- AvNumCits
- …
Data 3:
Not in tidy form
- station
- year
- month
- day
- TMAX
- TMIN
- PRCP
Statistical data
\[X = \left[ \begin{array}{rrrr} X_{~1} & X_{~2} & ... & X_{~p} \end{array} \right] \\ = \left[ \begin{array}{rrrr} X_{~11} & X_{~12} & ... & X_{~1p} \\ X_{~21} & X_{~22} & ... & X_{~2p} \\ \vdots & \vdots & \ddots& \vdots \\ X_{~n1} & X_{~n2} & ... & X_{~np} \end{array} \right]\]
- This is tidy data!
- You might also make assumptions about the distribution of each variable, e.g. \(X_{~1} \sim N(0,1), ~~X_{~2} \sim \text{Exp}(1) ...\)
Mapping
In ggplot2, the variables from tidy data are explicitly mapped to elements of the plot, using aesthetics.
Basic Mappings
xandyto plot points in a two-dimensional spacecolor,fillto render as a color scalesizemaps variable to size of objectshapemaps variable to different shapes
Depending on the geom different mappings are possible, xmin, xend, linetype, alpha, stroke, weight …
Facets
Variables are used to subset (or condition)
Layers
Different data can be mapped onto the same plot, eg observations, and means
Common plot descriptions as scripts
Example 1A
Example 1B
Example 2A
How are variables mapped to create this plot?
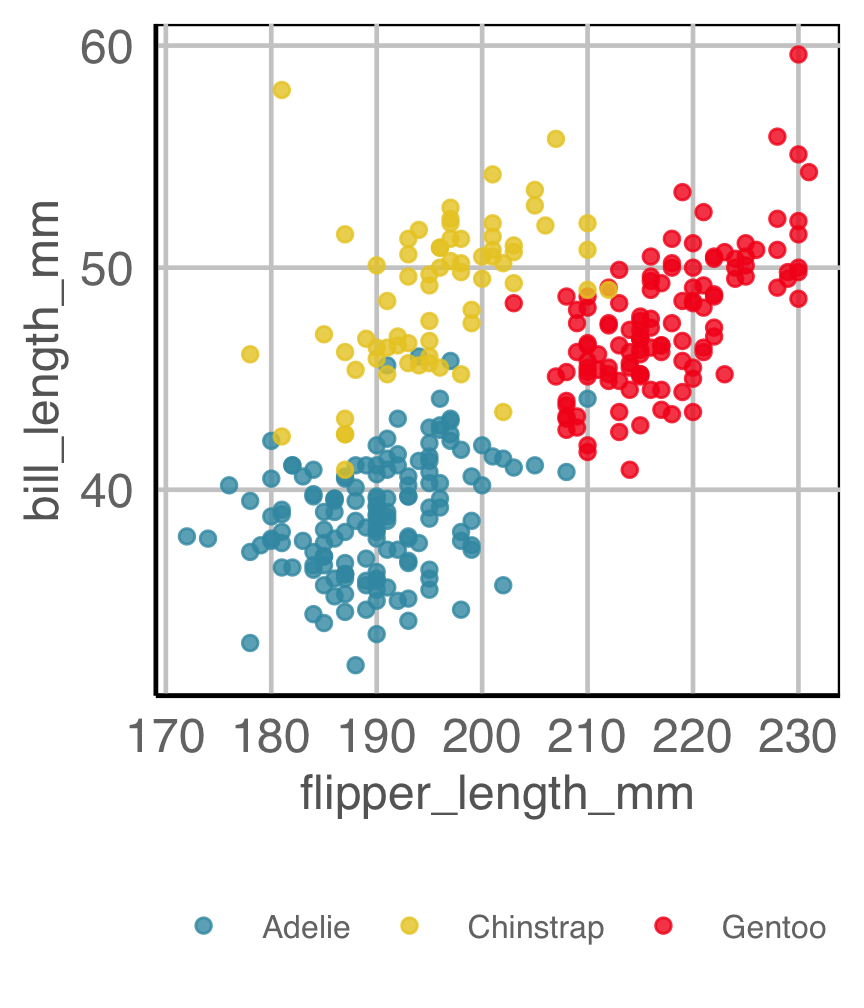
Code for penguins plot
Example 2B
How are variables mapped to create this plot?
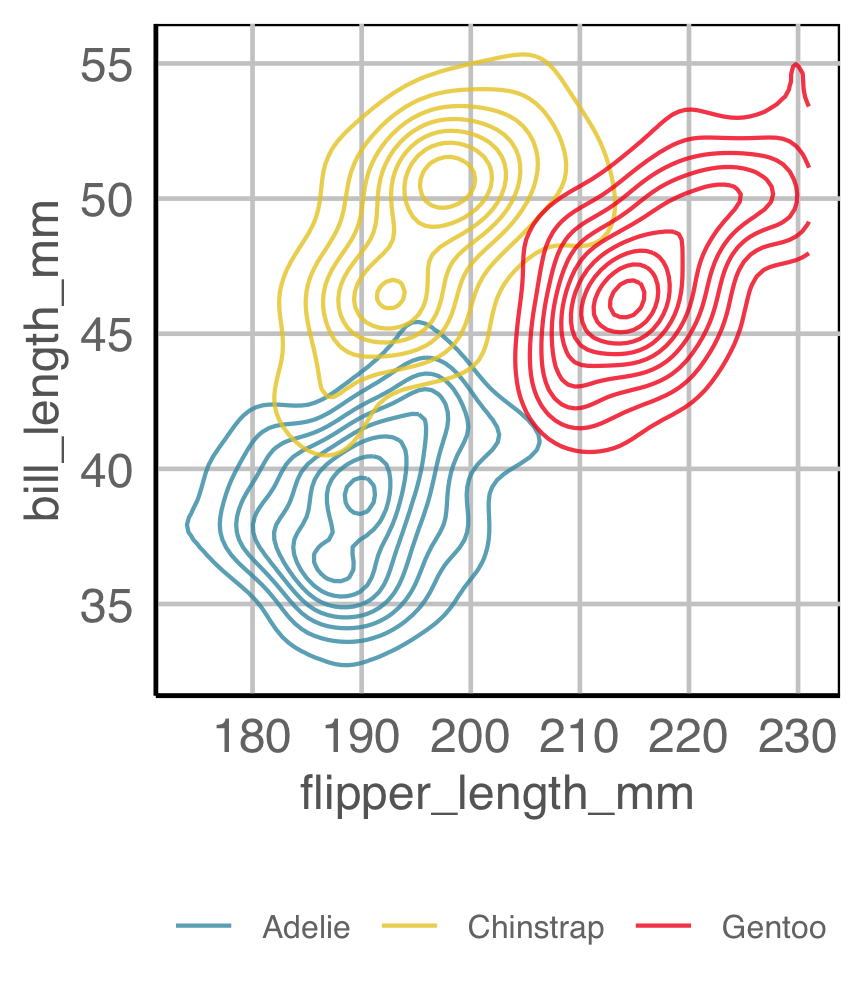
Code for penguins plot
Example 3 (1/5)
First get the data in tidy form
tb_aus_sa <- tb_aus |>
filter(year > 2012) |>
select(iso3, year,
newrel_f014:newrel_f65,
newrel_m014:newrel_m65) |>
pivot_longer(cols=newrel_f014:newrel_m65,
names_to = "sex_age",
values_to = "count") |>
filter(!is.na(count)) |>
separate(sex_age, into=c("stuff",
"sex_age")) |>
mutate(sex = str_sub(sex_age, 1, 1),
age = str_sub(sex_age, 2,
str_length(sex_age))) |>
mutate(age = case_when(
age == "014" ~ "0-14",
age == "1524" ~ "15-24",
age == "2534" ~ "25-34",
age == "3544" ~ "35-44",
age == "4554" ~ "45-54",
age == "5564" ~ "55-64",
age == "65" ~ "65")) |>
select(iso3, year, sex, age, count)Example 3 (2/5)
How many ways can we plot all three variables?
geom: bar + position (stack, dodge, fill)
aes:
- Var 1 to
x counttoy- Var 2 to
color - Var 3 to
facet
geom: point + smooth
aes:
- Var 1 to
x counttoy- Var 2 to
color - Var 3 to
facet
Example 3 (3/5)
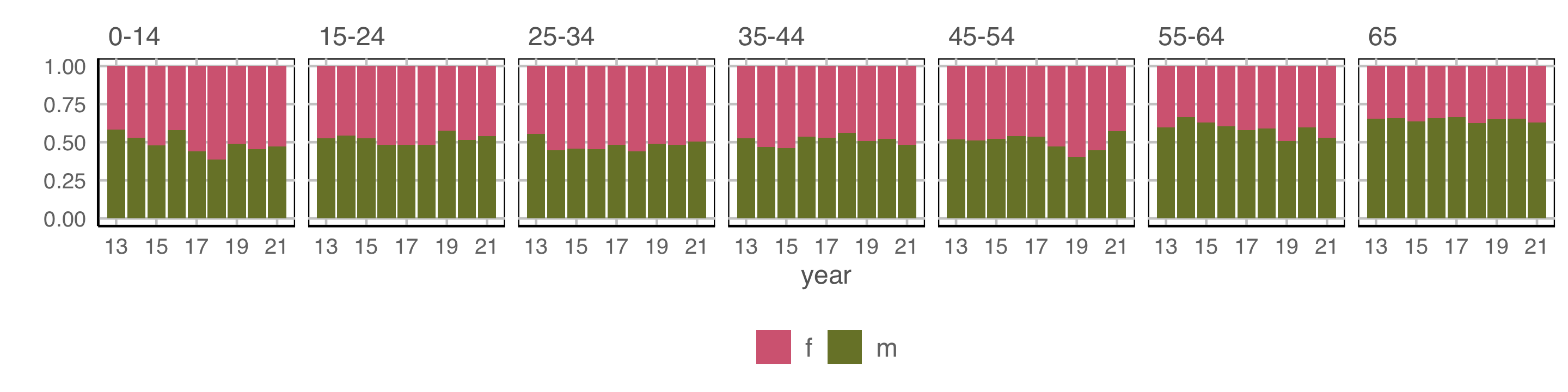
How are variables mapped to create this plot?
geom: bar/position=“fill”
year to x \(~~~~\) count to y \(~~~~\) fill to sex \(~~~~\) facet by age
Observations: Relatively equal proportions, with more incidence among males in older population. No clear temporal trend.
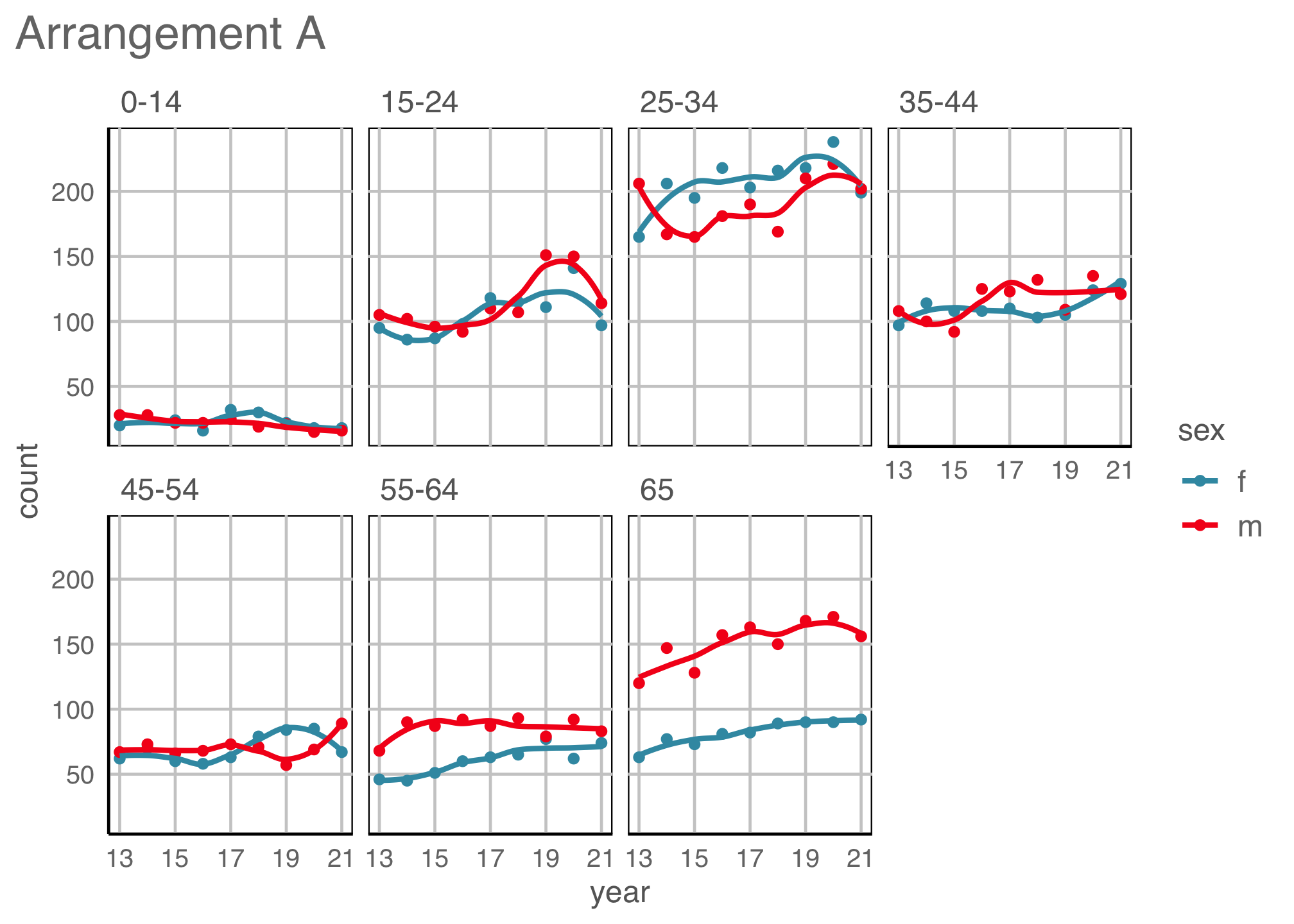
Example 3 (4/5)
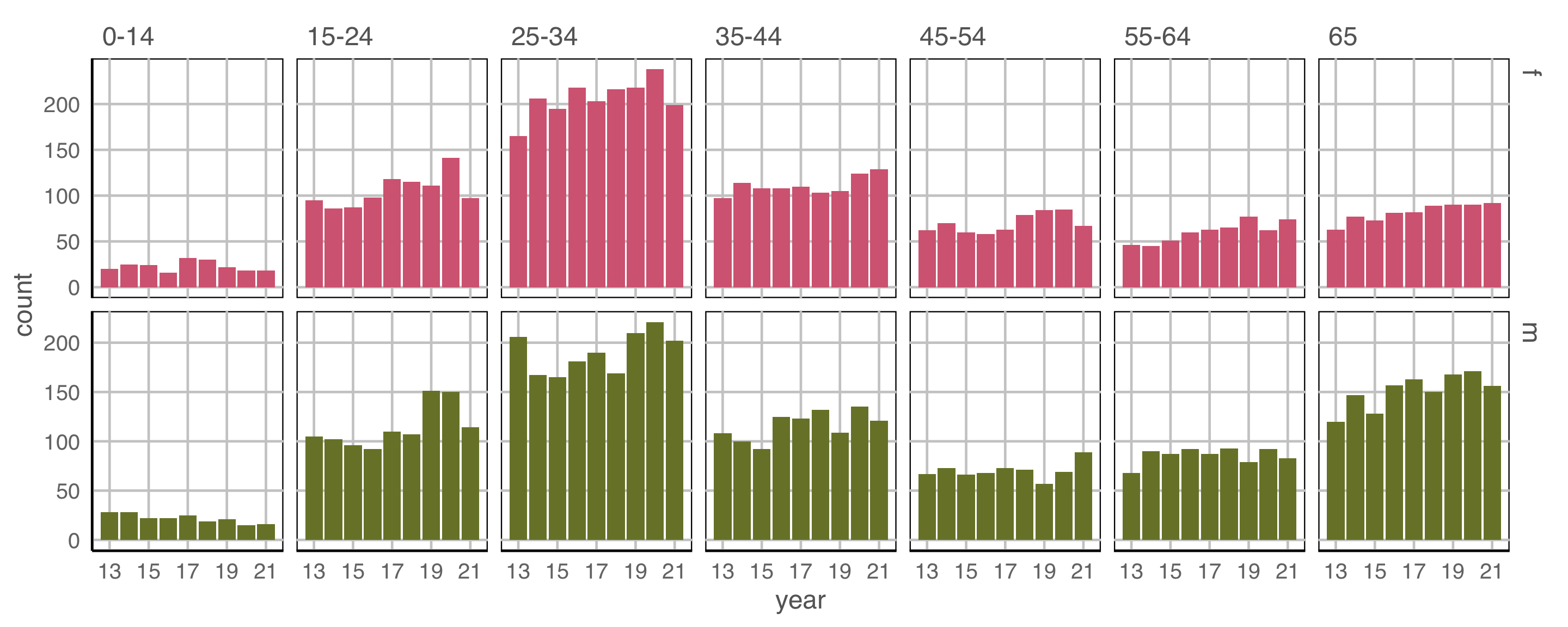
geom: bar
year to x \(~\) count to y \(~\) fill and facet to sex \(~\) facet by age
Incidence is higher among young adult groups, and older males.
Where’s the temporal trend?

Example 3 (5/5)
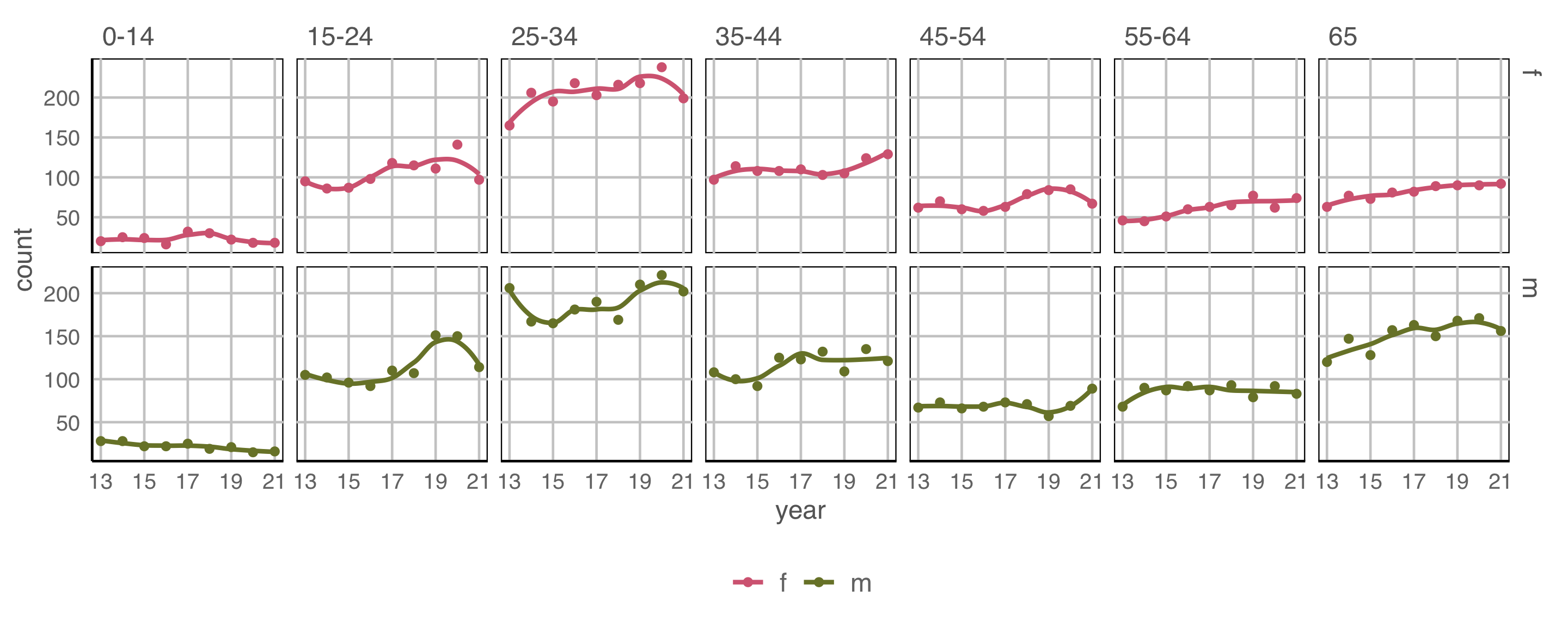
geom: point, smooth
year to x \(~\) count to y \(~\) colour and facet to sex \(~\) facet by age
Temporal trend is only present in some groups.

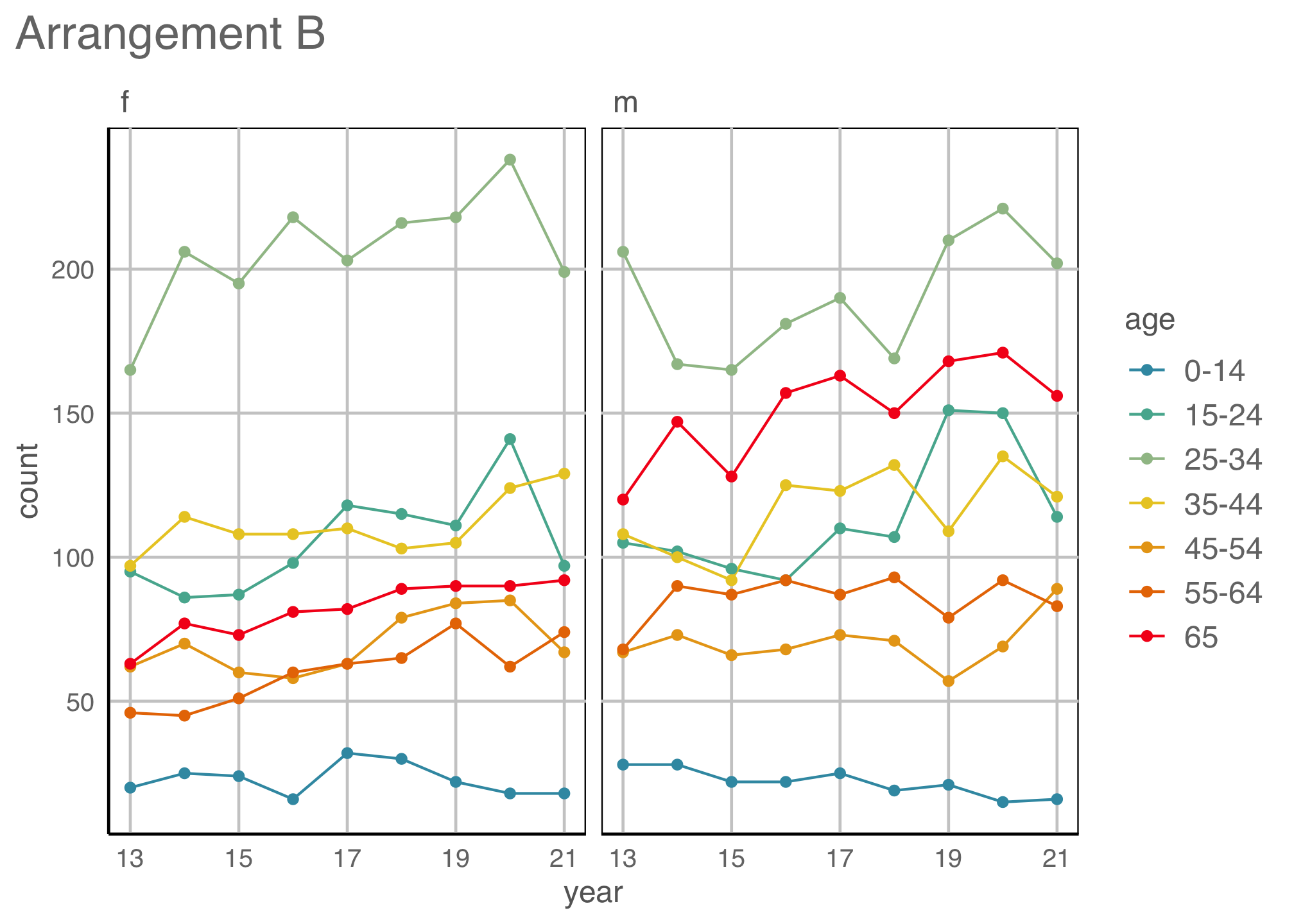
Tidy data to many plot descriptions
Re-arrangements
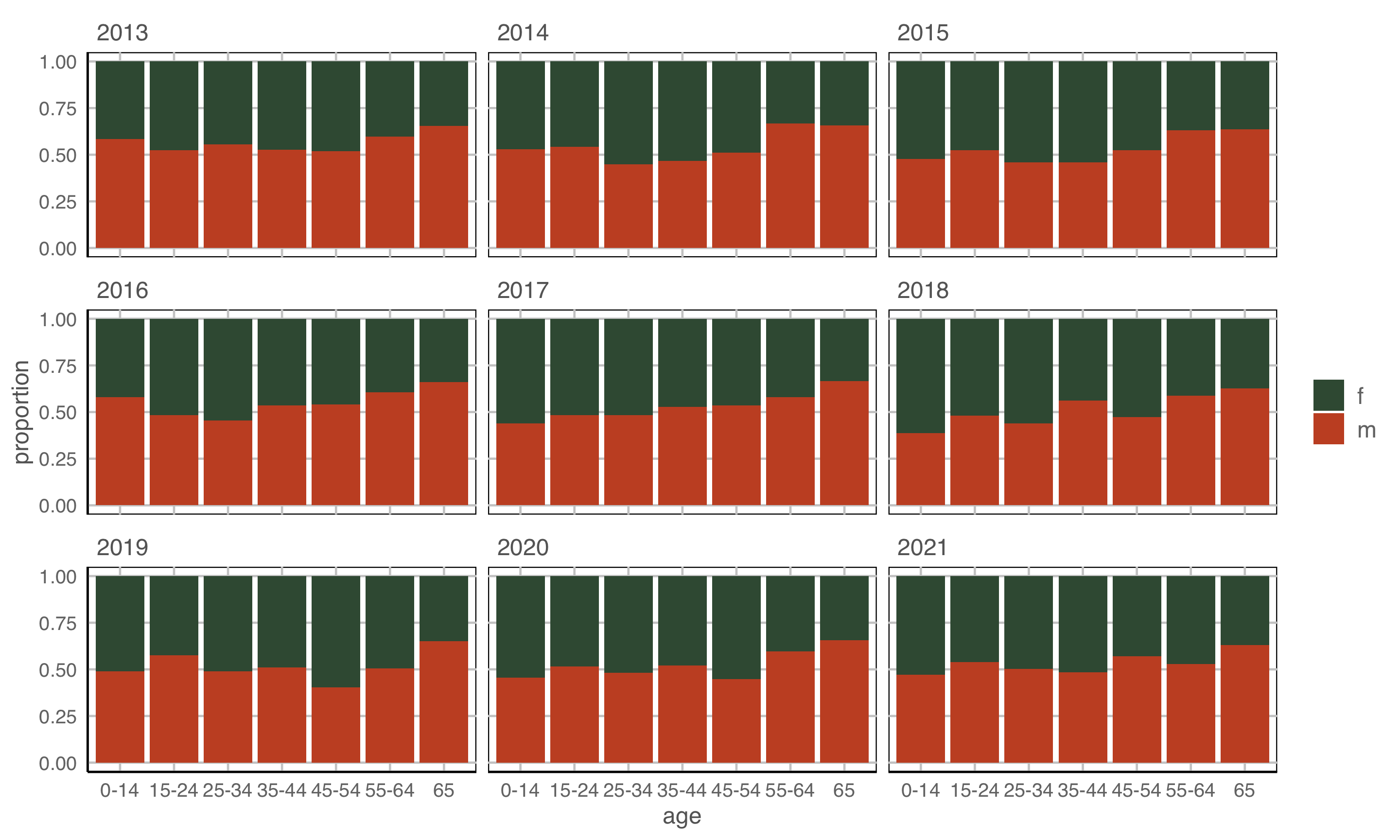
Scaling
This might have slipped under the radar, but different displays had some different scaling of the data:
Slides 6, 27, 28 (Why, Example 3 4,5/5) were constructed with
Why?
The emphasis was comparing difference in trend not magnitude of values.
Exercise
- Create two new plot arrangements of the Australian TB data. What do they put the focus on?
- Change the scaling on one of the plots, where it will change the appearance. How does the information change?
Do’s and don’ts
Plot descriptions
Use a new variable in a single data set - avoid multiple data sets (Tidy data principle)
GOOD
BAD
ggplot() +
geom_point(data = tb_aus,
aes(x=year, y=c_newinc),
colour="#F5191C") +
geom_point(data = tb_idn,
aes(x=year, y=c_newinc),
colour="#3B99B1") +
geom_smooth(data = tb_aus,
aes(x=year, y=c_newinc),
colour="#F5191C", se=F) +
geom_smooth(data = tb_idn,
aes(x=year, y=c_newinc),
colour="#3B99B1", se=F) Cognitive principles
Hierarchy of mappings
Cleveland and McGill (1984)
Illustrations made by Emi Tanaka
Hierarchy of mappings
- Position - common scale (BEST)
- Position - nonaligned scale
- Length, direction, angle
- Area
- Volume, curvature
- Shading, color (WORST)
- scatterplot, barchart
- side-by-side boxplot, stacked barchart
- piechart, rose plot, gauge plot, donut, wind direction map, starplot
- treemap, bubble chart, mosaicplot
- chernoff face
- choropleth map
Proximity
Place elements that you want to compare close to each other. If there are multiple comparisons to make, you need to decide which one is most important.
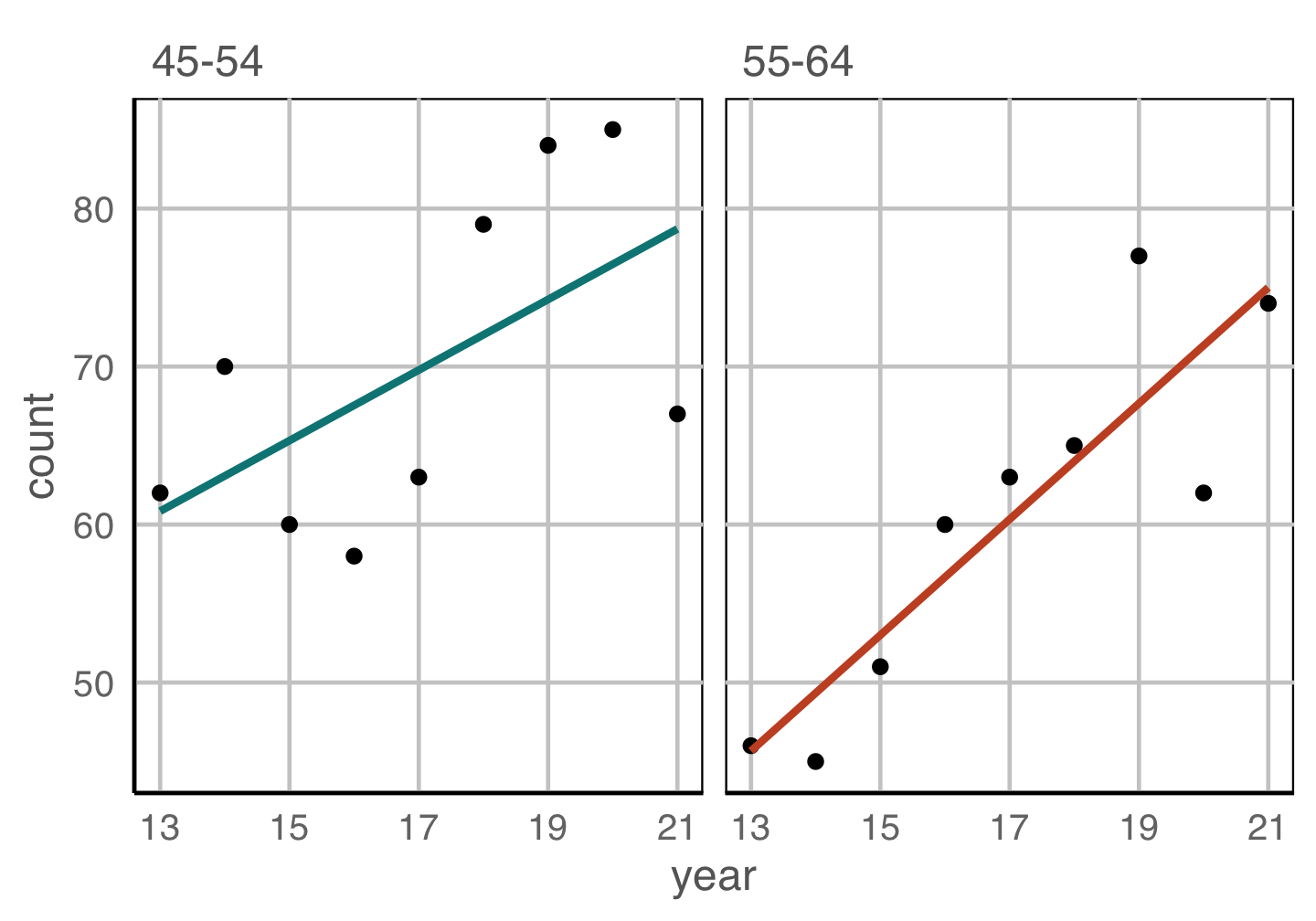
Change blindness
Making comparisons across plots requires the eye to jump from one focal point to another. It may result in not noticing differences.
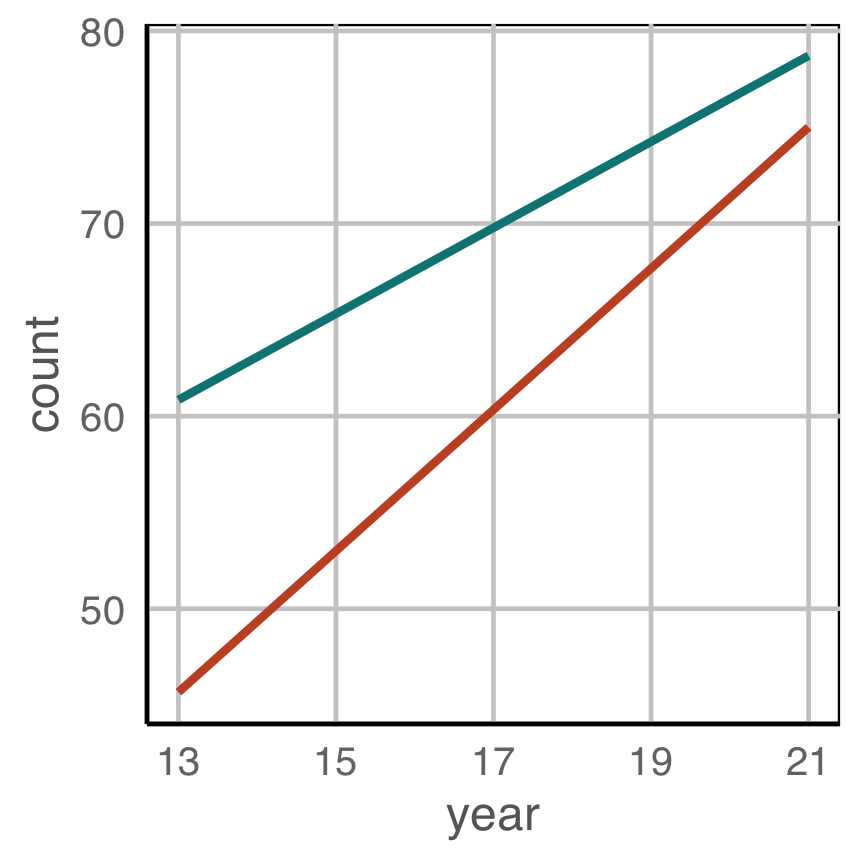
Exercise
Take the following plot, and make it more difficult to read.
Think about what is it you learn from the plot, and how
- changing the mapping,
- using colour, or
- the geom type
might change what you learn.
Take-aways
- Organise your data so that variables are clear
- Clearly map your variables to elements of the plot
- It’s ok to make many plots of the same variables
- Use proximity effectively
Long exercise
This data is downloaded from ABS Census Datapacks. For this data the goal is to fix the code below for plotting the distribution of household income across Victorian LGAs.
- What are the disadvantages of using the code as is?
- Rearrange the data into tidy form.
- Plot facetted histograms of the distribution of counts by household type.
- Plot the data another way, eg density, boxplot.
- What do you learn about difference in distributions between family and non-family households?
Solution to long exercise and more
Code for answer
file = "data/2021Census_G33_VIC_LGA.csv"
hh_income <- read_csv(file)
hh_tidy <- hh_income |>
select(LGA_CODE_2021,
Tot_Family_households,
Tot_Non_family_households) |>
pivot_longer(cols=contains("Tot"),
names_to="hh_type",
values_to="count") |>
mutate(hh_type = str_remove(hh_type, "Tot_")) |>
mutate(hh_type = str_remove(hh_type, "_households")) |>
mutate(hh_type = str_remove(hh_type, "_family"))
ggplot(hh_tidy, aes(x=count)) +
geom_histogram() +
facet_wrap(~hh_type, ncol=1)
ggplot(hh_tidy, aes(x=count,
colour=hh_type,
fill=hh_type)) +
geom_density(alpha=0.5) +
scale_color_discrete_divergingx() +
scale_fill_discrete_divergingx()
ggplot(hh_tidy, aes(x=hh_type,
y=count)) +
geom_boxplot()
ggplot(hh_tidy, aes(x=hh_type,
y=count)) +
geom_quasirandom() Code for proportions
hhi_tidy <- hh_income |>
select(LGA_CODE_2021,
contains("_Tot"), -Tot_Tot) |>
pivot_longer(cols=contains("Tot"),
names_to="income_cat",
values_to="count") |>
mutate(income_cat = str_remove(income_cat, "_Tot")) |>
mutate(income_cat = str_remove(income_cat, "HI_")) |>
mutate(income_cat = str_remove(income_cat, "_Nil_income")) |>
mutate(income_cat = str_remove(income_cat, "_income_stated")) |>
mutate(income_cat = str_remove(income_cat, "_incomes_not_stated")) |>
group_by(income_cat) |>
mutate(prop = count/sum(count)) |>
dplyr::filter(!(income_cat %in%
c("Partial", "All", "Negative"))) |>
separate(income_cat, into=c("cmin", "cmax")) |>
mutate(cmax = str_replace(cmax, "more", "5000")) |>
mutate(income = (as.numeric(cmin) +
as.numeric(cmax))/2) |>
select(-cmin, cmax)
hhi_tidy |>
dplyr::filter(LGA_CODE_2021 %in%
sample(unique(hhi_tidy$LGA_CODE_2021), 8)) |>
ggplot(aes(x=income, y=count)) +
facet_wrap(~LGA_CODE_2021, ncol=4) +
geom_line()End of session 1

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.