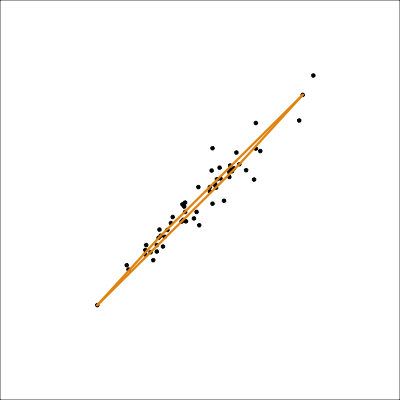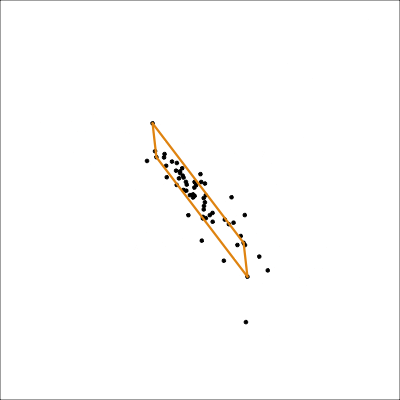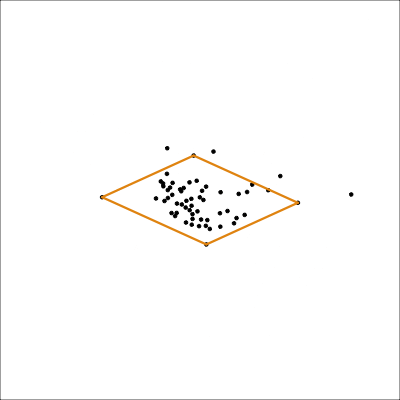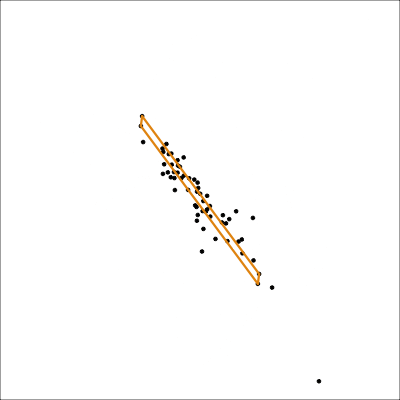| time | topic |
|---|---|
| 1:30-1:40 | Introduction: What is high-dimensional data, why visualise and overview of methods |
| 1:50-2:10 | Basics of linear projections, and recognising high-d structure |
| 2:10-2:30 | Effectively reducing your data dimension, in association with non-linear dimension reduction |
| 2:30-3:00 | BREAK |
Visualising High-dimensional Data with R
Session 1
Session 1
Outline
Introduction
What is high-dimensional space?
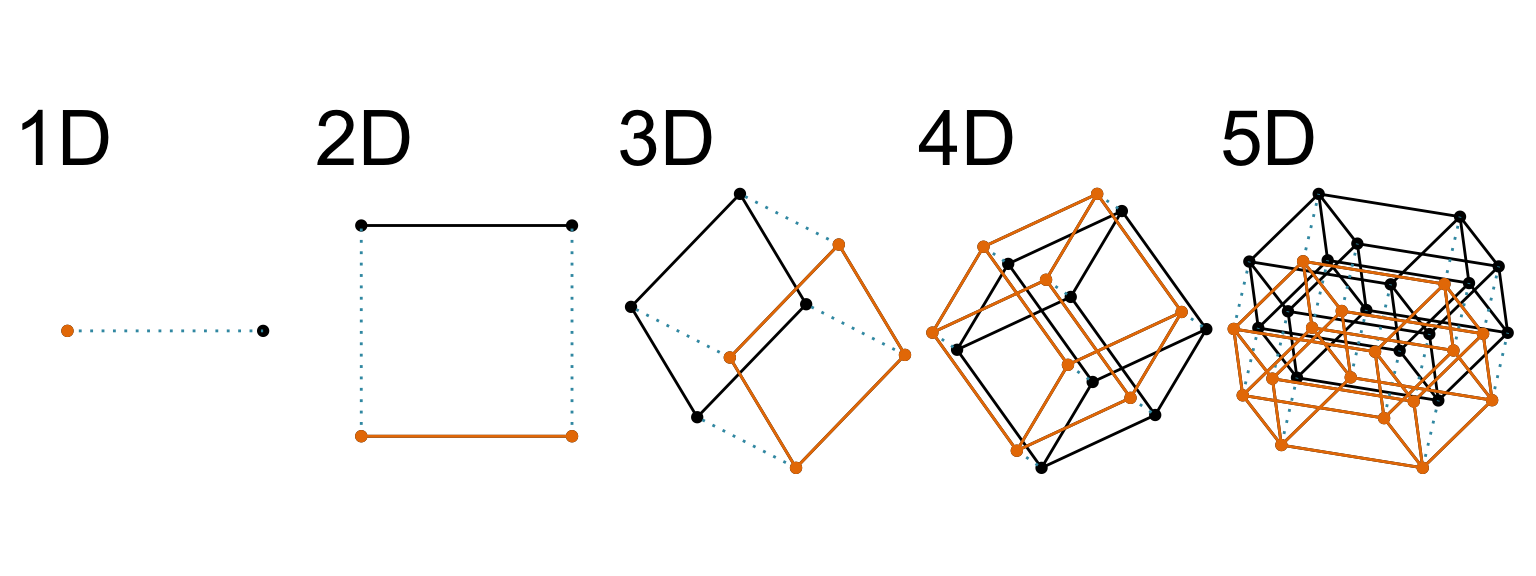
Increasing dimension adds an additional orthogonal axis.
If you want more high-dimensional shapes there is an R package, geozoo, which will generate cubes, spheres, simplices, mobius strips, torii, boy surface, klein bottles, cones, various polytopes, …
And read or watch Flatland: A Romance of Many Dimensions (1884) Edwin Abbott.
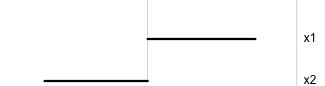
Notation: Data
\[\begin{eqnarray*} X_{~n\times p} = [X_{~1}~X_{~2}~\dots~X_{~p}]_{~n\times p} = \left[ \begin{array}{cccc} x_{~11} & x_{~12} & \dots & x_{~1p} \\ x_{~21} & x_{~22} & \dots & x_{~2p}\\ \vdots & \vdots & & \vdots \\ x_{~n1} & x_{~n2} & \dots & x_{~np} \end{array} \right]_{~n\times p} \end{eqnarray*}\]
Notation: Projection
\[\begin{eqnarray*} A_{~p\times d} = \left[ \begin{array}{cccc} a_{~11} & a_{~12} & \dots & a_{~1d} \\ a_{~21} & a_{~22} & \dots & a_{~2d}\\ \vdots & \vdots & & \vdots \\ a_{~p1} & a_{~p2} & \dots & a_{~pd} \end{array} \right]_{~p\times d} \end{eqnarray*}\]
Notation: Projected data
\[\begin{eqnarray*} Y_{~n\times d} = XA = \left[ \begin{array}{cccc} y_{~11} & y_{~12} & \dots & y_{~1d} \\ y_{~21} & y_{~22} & \dots & y_{~2d}\\ \vdots & \vdots & & \vdots \\ y_{~n1} & y_{~n2} & \dots & y_{~nd} \end{array} \right]_{~n\times d} \end{eqnarray*}\]
Why? (1/2)
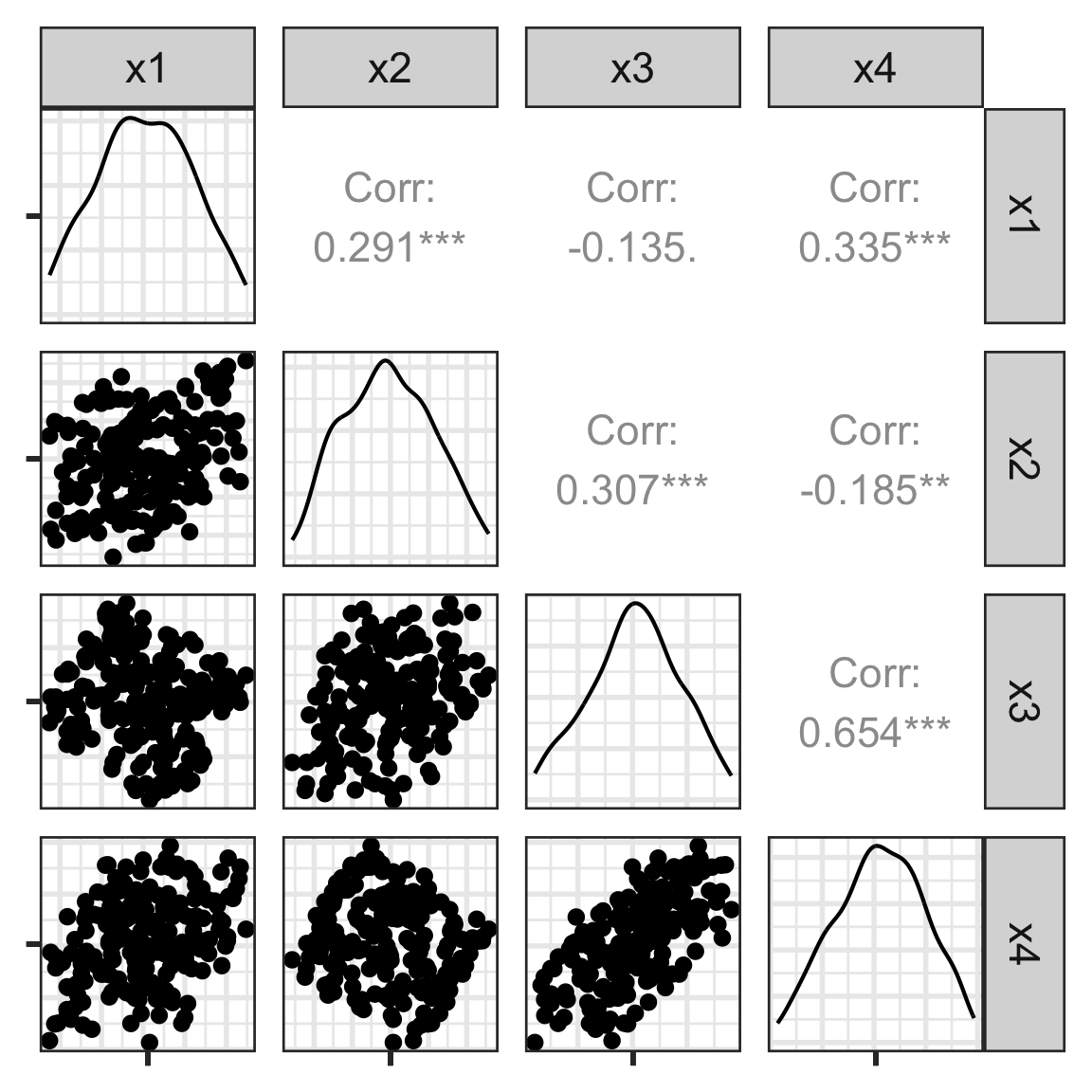
Scatterplot matrix
Here, we see linear association, clumping and clustering, potentially some outliers.

Why? (2/2)

There is an outlier in the data on the right, like the one in the left, but it is hidden in a combination of variables. It’s not visible in any pair of variables.
And help to see the data as a whole
To avoid misinterpretation …
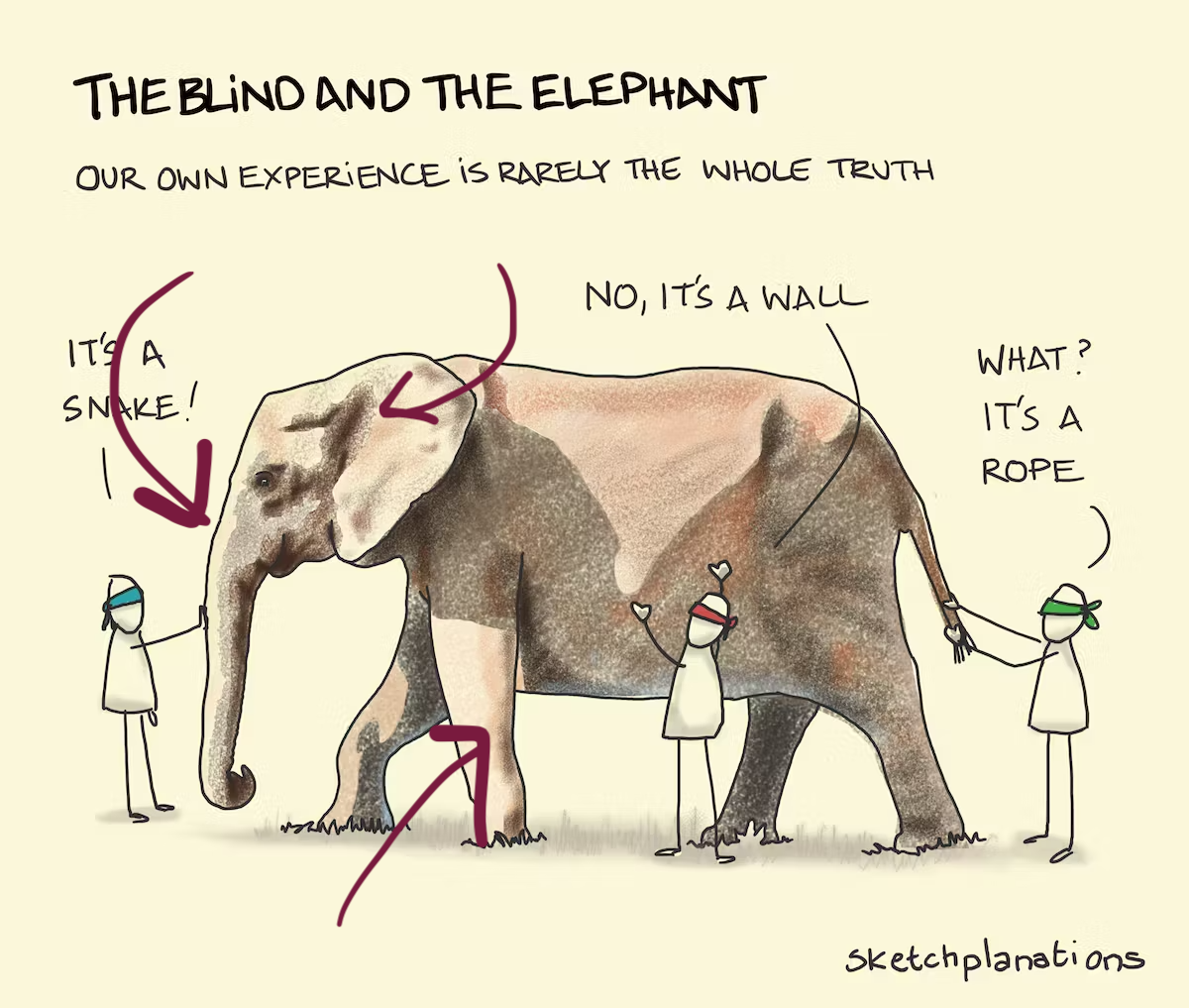
… see the bigger picture!
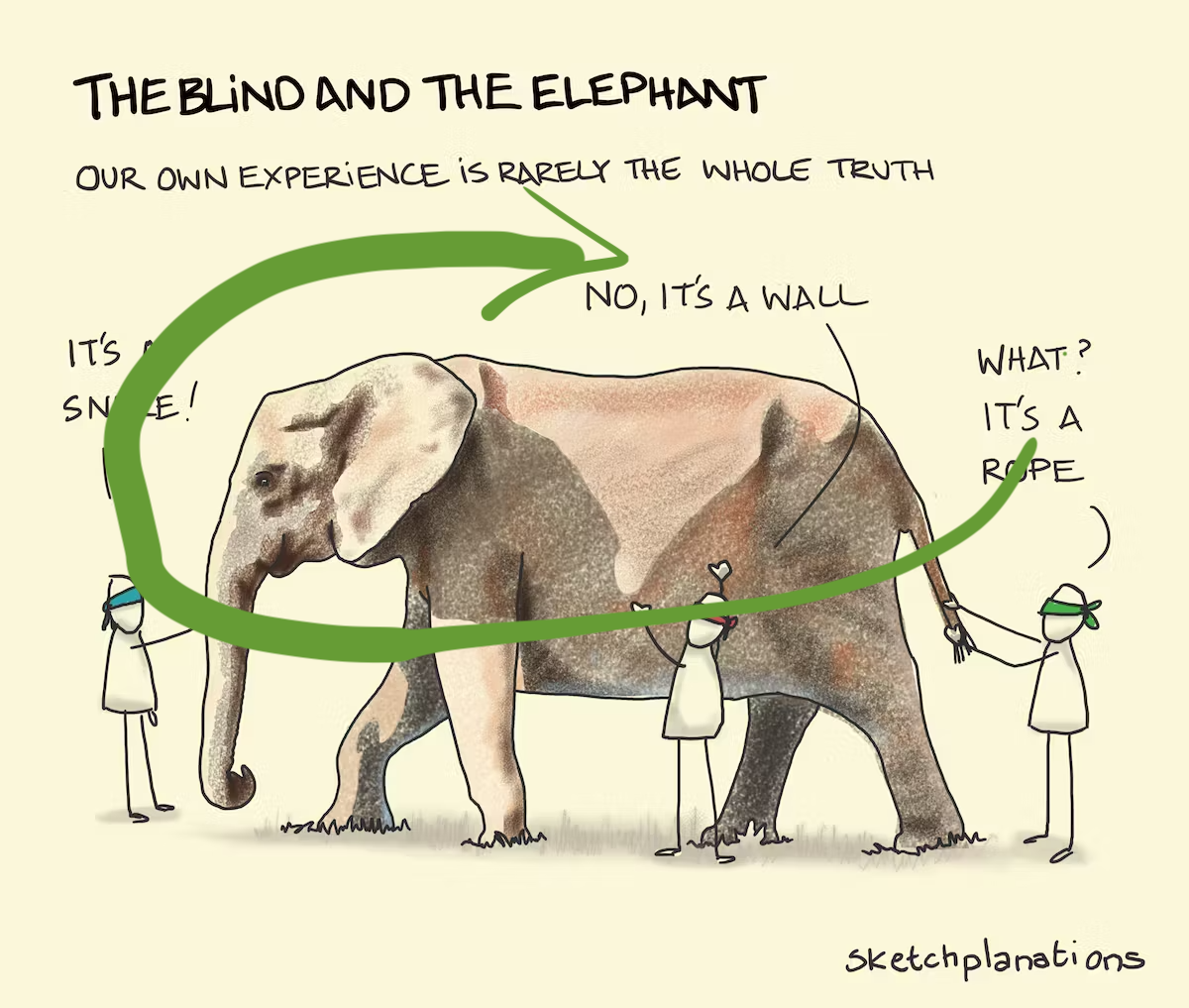
Image: Sketchplanations.
Tours of linear projections
Data is 2D: \(~~p=2\)
Projection is 1D: \(~~d=1\)\[\begin{eqnarray*} A_{~2\times 1} = \left[ \begin{array}{c} a_{~11} \\ a_{~21}\\ \end{array} \right]_{~2\times 1} \end{eqnarray*}\]
Notice that the values of \(A\) change between (-1, 1). All possible values being shown during the tour.
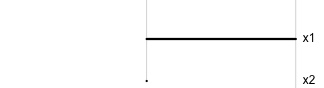
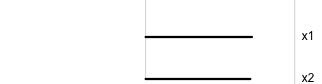
\[\begin{eqnarray*} A = \left[ \begin{array}{c} 1 \\ 0\\ \end{array} \right] ~~~~~~~~~~~~~~~~ A = \left[ \begin{array}{c} 0.7 \\ 0.7\\ \end{array} \right] ~~~~~~~~~~~~~~~~ A = \left[ \begin{array}{c} 0.7 \\ -0.7\\ \end{array} \right] \end{eqnarray*}\]
watching the 1D shadows we can see:
- unimodality
- bimodality, there are two clusters.
What does the 2D data look like? Can you sketch it?
Tours of linear projections

⟵
The 2D data

Tours of linear projections
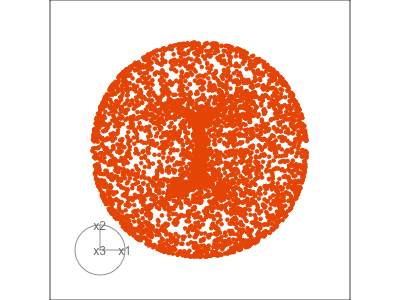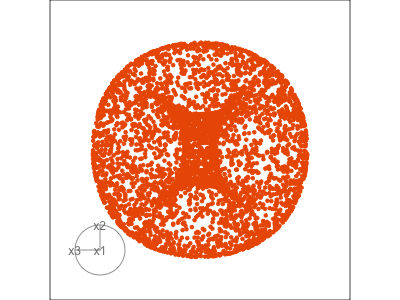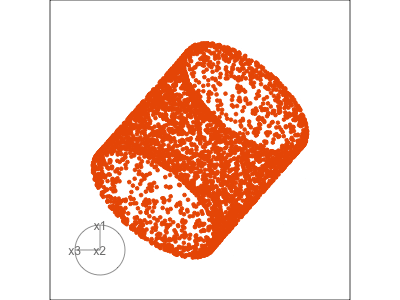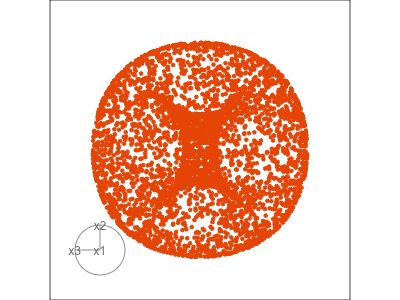
Data is 3D: \(p=3\)
Projection is 2D: \(d=2\)
\[\begin{eqnarray*} A_{~3\times 2} = \left[ \begin{array}{cc} a_{~11} & a_{~12} \\ a_{~21} & a_{~22}\\ a_{~31} & a_{~32}\\ \end{array} \right]_{~3\times 2} \end{eqnarray*}\]
Notice that the values of \(A\) change between (-1, 1). All possible values being shown during the tour.
See:
- circular shapes
- some transparency, reveals middle
- hole in in some projections
- no clustering
Tours of linear projections
Data is 4D: \(p=4\)
Projection is 2D: \(d=2\)
\[\begin{eqnarray*} A_{~4\times 2} = \left[ \begin{array}{cc} a_{~11} & a_{~12} \\ a_{~21} & a_{~22}\\ a_{~31} & a_{~32}\\ a_{~41} & a_{~42}\\ \end{array} \right]_{~4\times 2} \end{eqnarray*}\]
How many clusters do you see?
- three, right?
- one separated, and two very close,
- and they each have an elliptical shape.
- do you also see an outlier or two?
Intuitively, tours are like …
Anomaly is no longer hidden

How to use a tour in R
This is a basic tour, which will run in your RStudio plot window.
You can specifically guide the tour choice of projections using
How to save a tour
Your turn
Use a grand tour on the data set c1 in the mulgar package. What shapes do you see?
Have a look at c3 or c7 also. How are the structures different.
05:00 Dimension reduction
What is dimensionality?

When an axis extends out of a direction where the points are collapsed, it means that this variable is partially responsible for the reduced dimension.
In high-dimensions
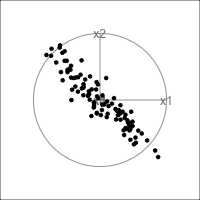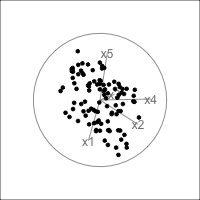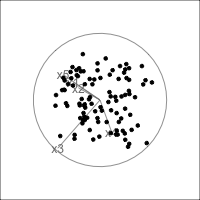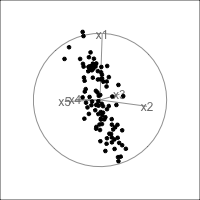
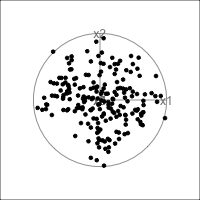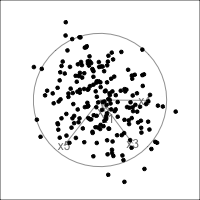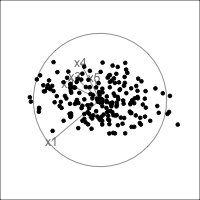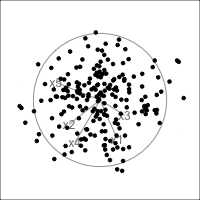
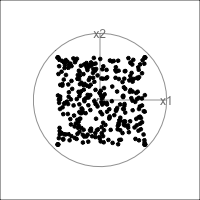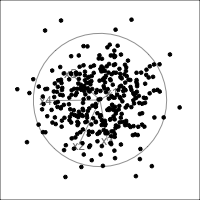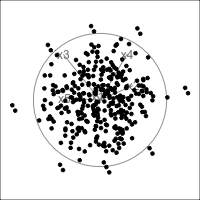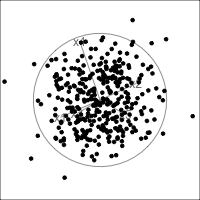
Principal component analysis (PCA) will detect these dimensionalities.
Some data is basically univariate

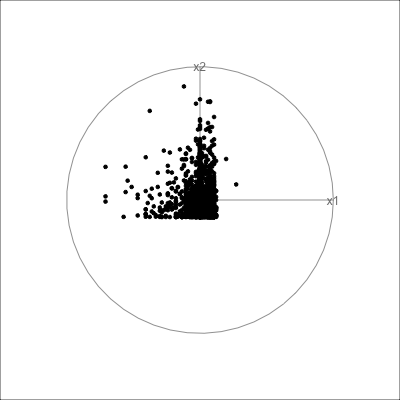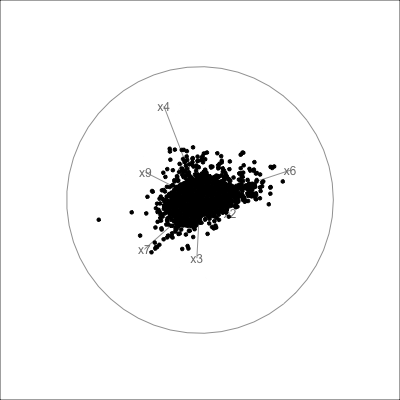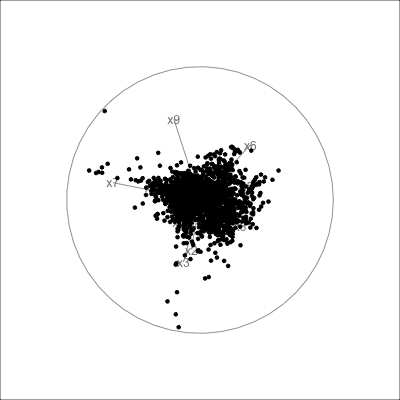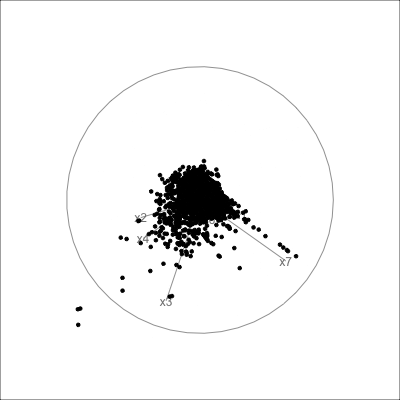
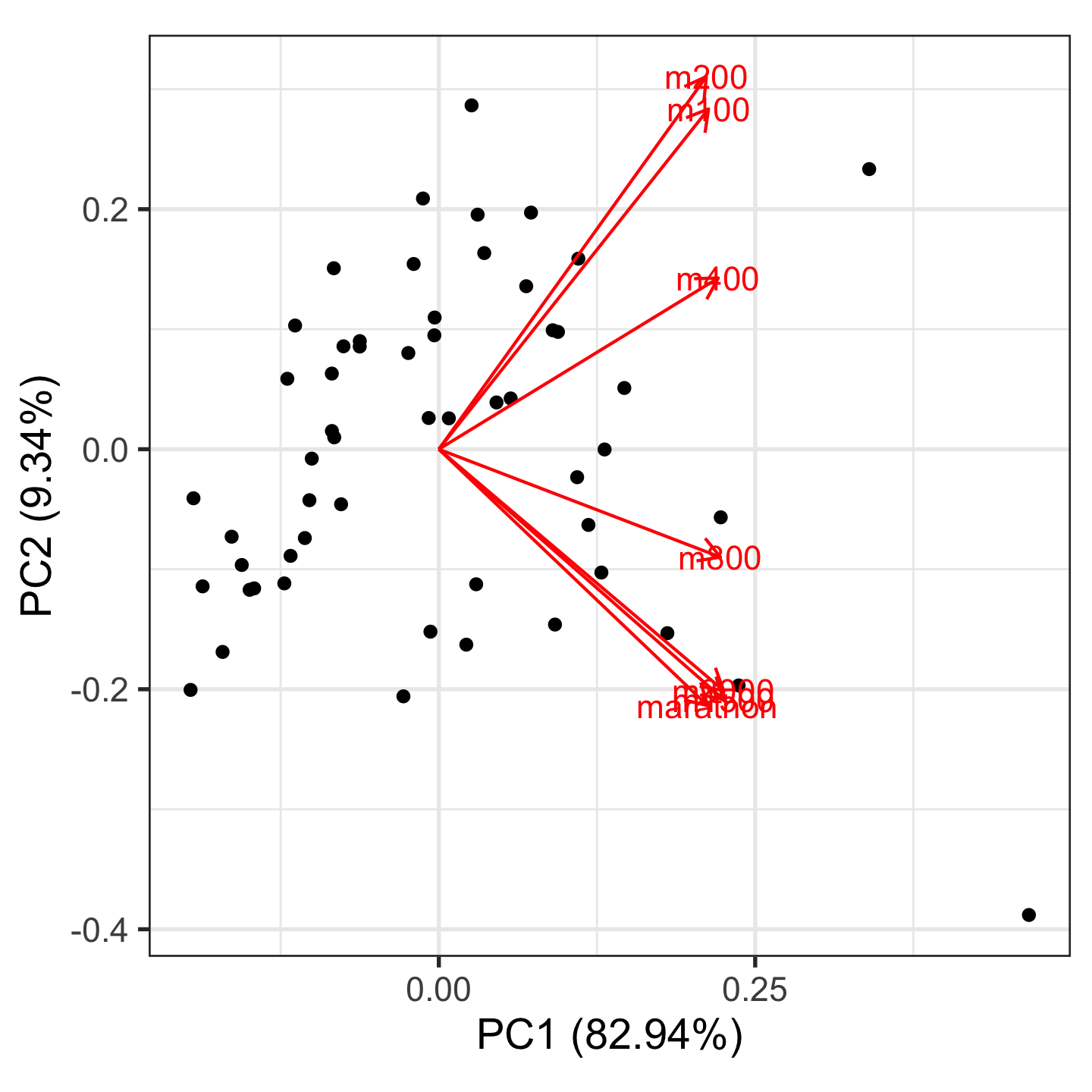
Example: womens’ track records (1/3)

Source: Johnson and Wichern, Applied multivariate analysis
Example: PCA summary (2/3)
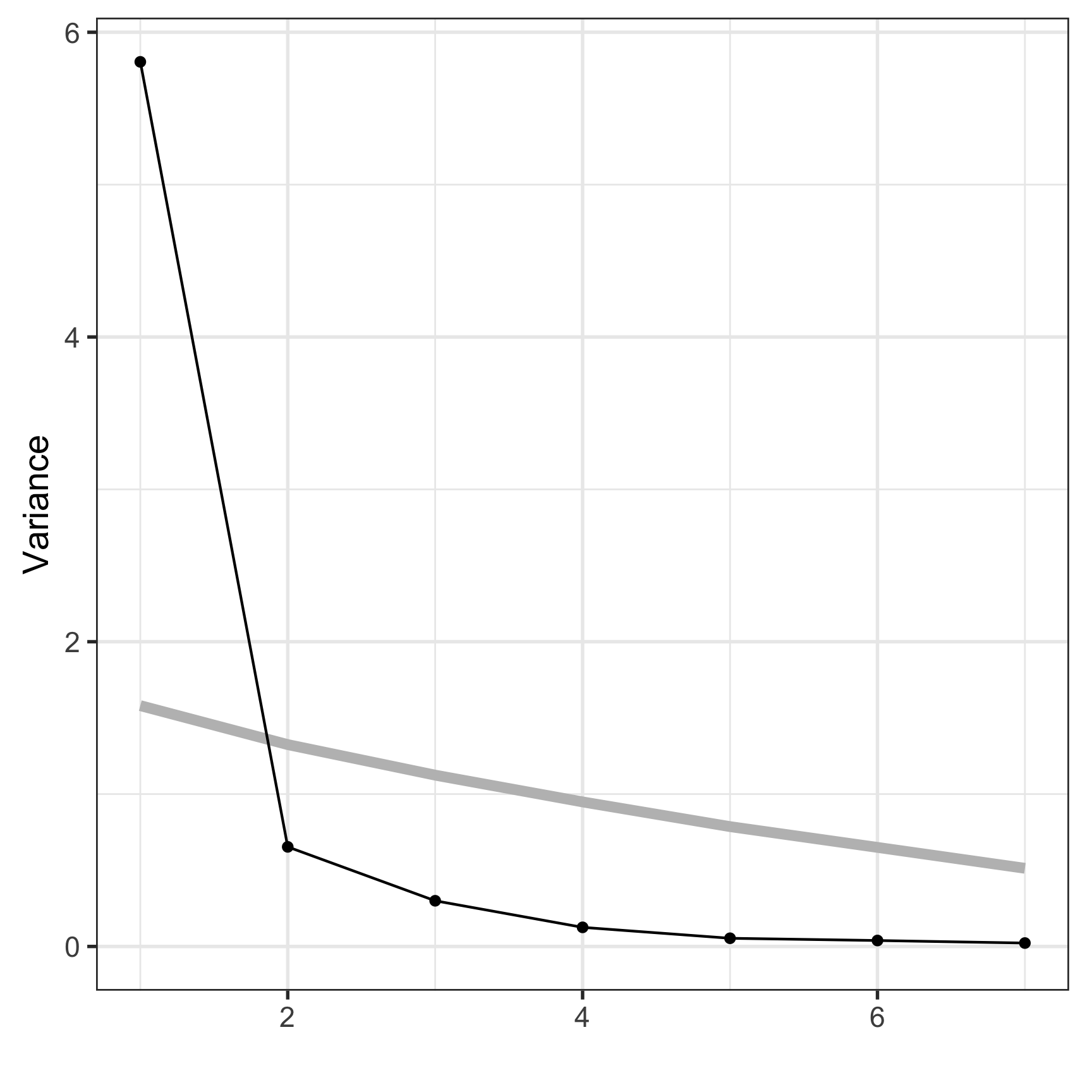
Variances/eigenvalues
[1] 5.806 0.654 0.300 0.125 0.054 0.039 0.022Component coefficients
PC1 PC2 PC3 PC4
m100 0.37 0.49 -0.286 0.319
m200 0.37 0.54 -0.230 -0.083
m400 0.38 0.25 0.515 -0.347
m800 0.38 -0.16 0.585 -0.042
m1500 0.39 -0.36 0.013 0.430
m3000 0.39 -0.35 -0.153 0.363
marathon 0.37 -0.37 -0.484 -0.672How many PCs?
Example: Visualise (3/3)
Key conceptual points
- Avoid misinterpretation, by using your high-dimensional visualisation skills to look at the data as a whole.
- Examine model fit by by examining the model overlaid on the data, model-in-the-data-space. (Wickham et al (2015) Removing the Blindfold)
End of session 1

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.