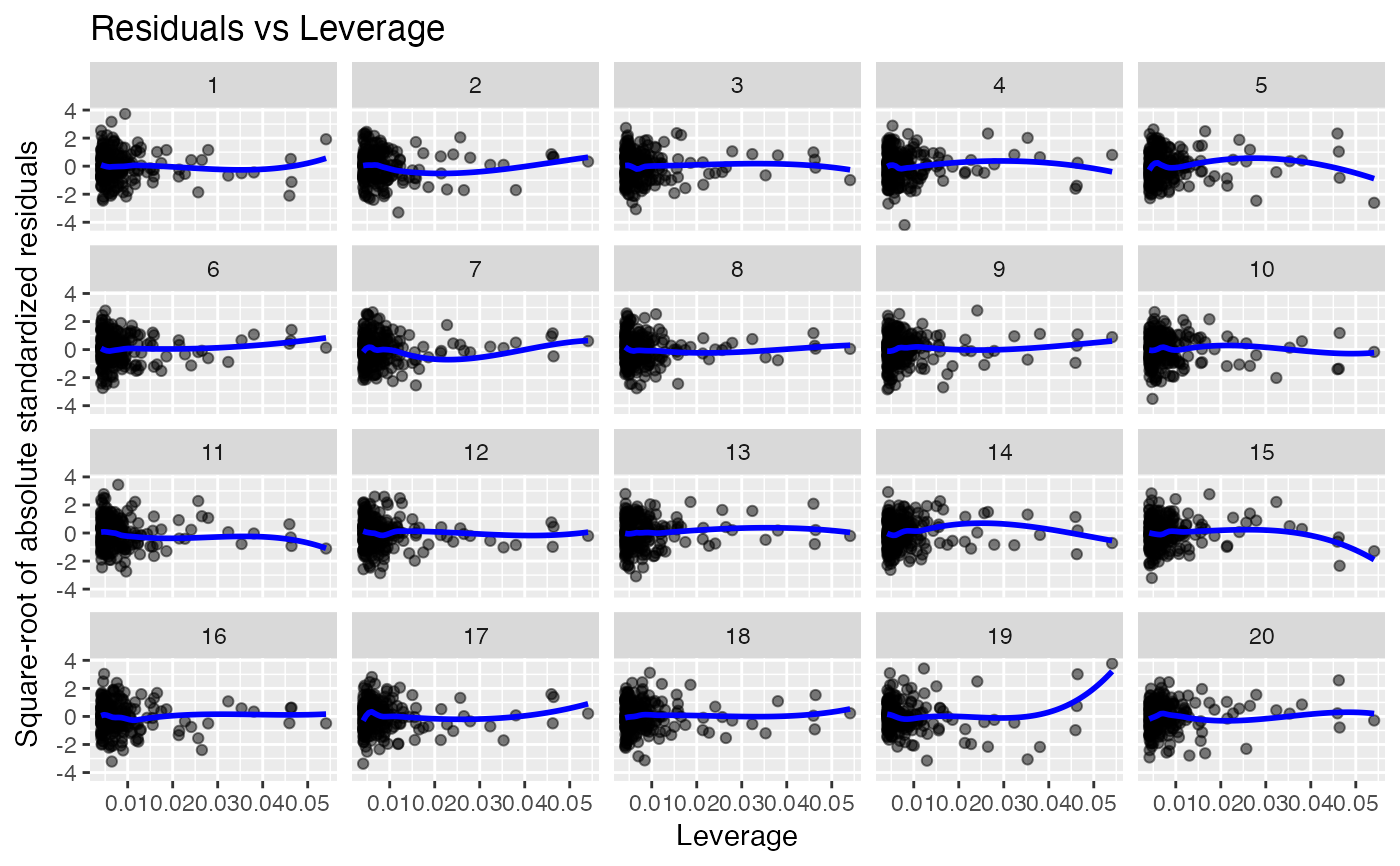
Compare residual plots of a fitted model to plots of null residuals.
Source:R/quick_plots.R
lineup_residuals.RdThis function is used to quickly create lineup version of the residual
plots created by plot.lm and ggfortify::autoplot.lm; see Details for
descriptions of these plots.
In the lineup protocol the plot of the real data is embedded amongst a field of
plots of data generated to be consistent with some null hypothesis.
If the observer can pick the real data as different from the others, this
lends weight to the statistical significance of the structure in the plot.
The protocol is described in Buja et al. (2009).
lineup_residuals(
model,
type = 1,
method = "rotate",
color_points = "black",
color_trends = "blue",
color_lines = "brown3",
alpha_points = 0.5,
...
)Arguments
- model
a model object fitted using
lm.- type
type of plot: 1 = residuals vs fitted, 2 = normal Q-Q, 3 = scale-location, 4 = residuals vs leverage.
- method
method for generating null residuals. Built in methods 'rotate', 'perm', 'pboot' and 'boot' are defined by
resid_rotate,resid_perm,resid_pbootandresid_bootrespectively. 'pboot' is always used for plots of type 2.- color_points
the color used for points in the plot. Can be a name or a color HEX code.
- color_trends
the color used for trend curves in the plot.
- color_lines
the color used for reference lines in the plot.
- alpha_points
the alpha (opacity) used for points in the plot (between 0 and 1, where 1 is opaque).
- ...
other arguments passed onto
method.
Value
a ggplot
Details
Four types of plots are available:
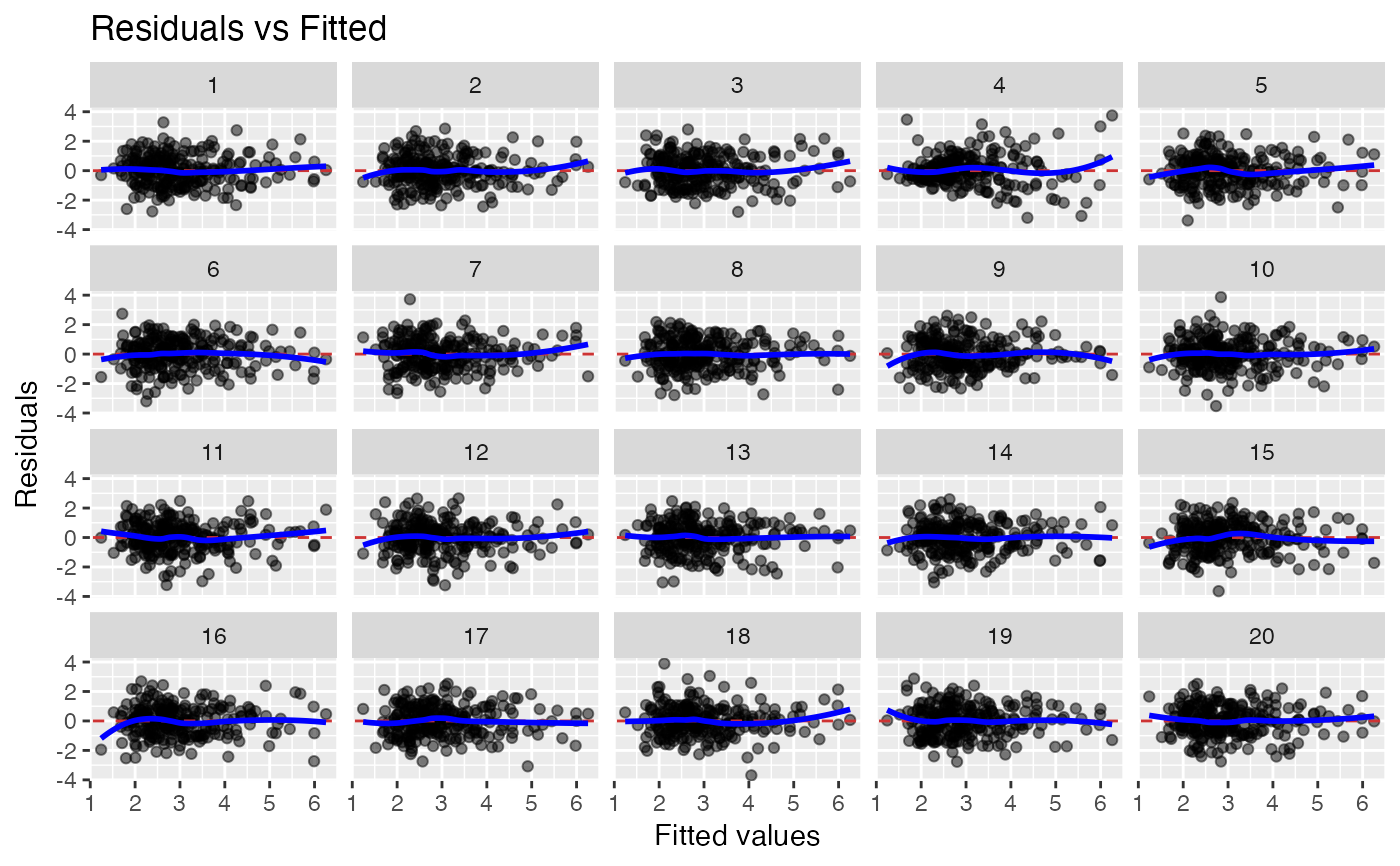
Residual vs fitted. Null hypothesis: variable is linear combination of predictors.
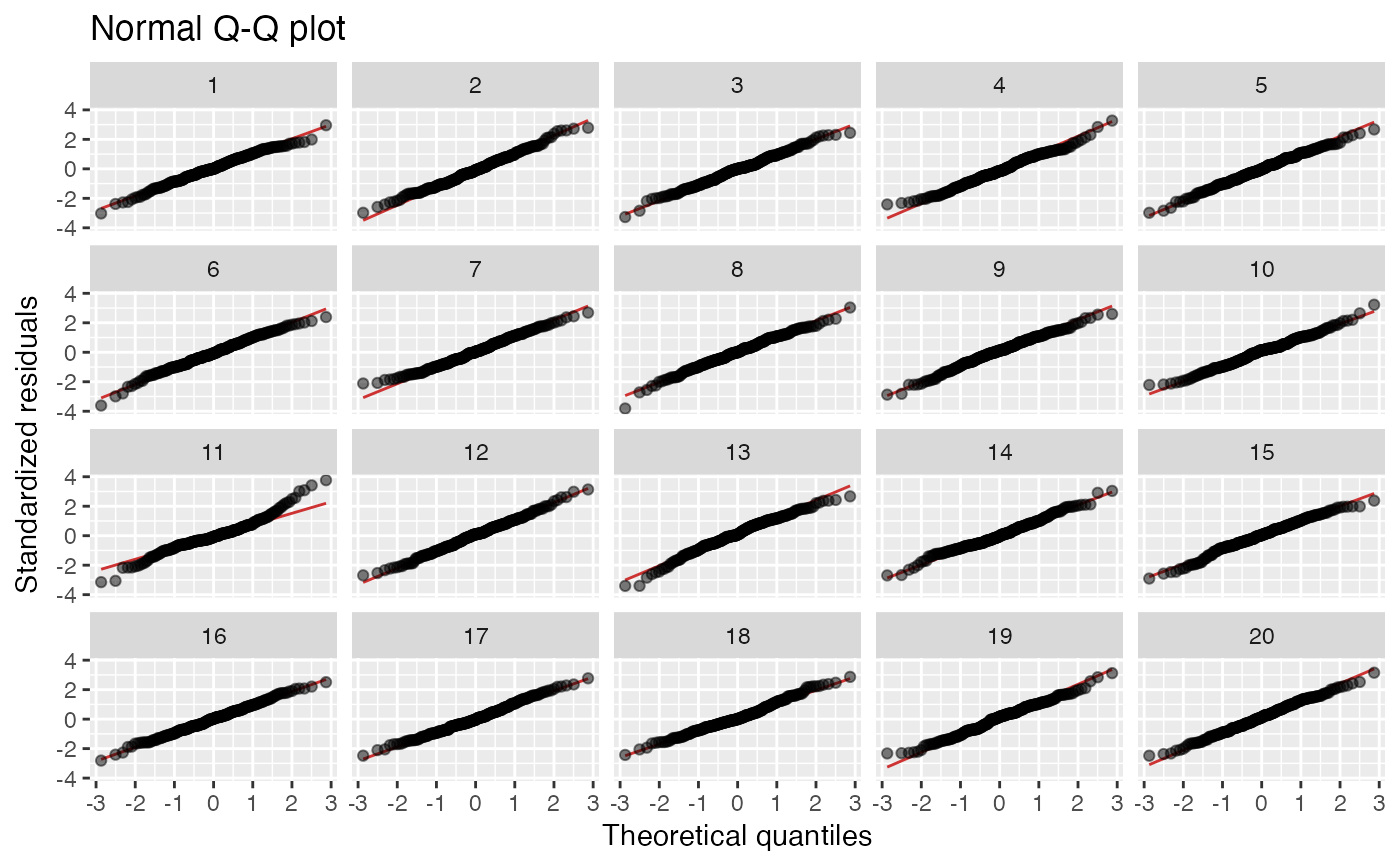
Normal Q-Q plot. Null hypothesis: errors are normal. Always uses
method = "pboot"to generate residuals under the null hypothesis.Scale-location. Null hypothesis: errors are homoscedastic.
Residuals vs leverage. Used to identify points with high residuals and high leverage, which are likely to have a strong influence on the model fit.
19 null datasets are plotted together the the true data (randomly
positioned). If you pick the real data as being noticeably different, then
you have formally established that it is different to with p-value 0.05.
Run the decrypt message printed in the R Console to see which
plot represents the true data.
If the null hypothesis in the type 1 plot is violated, consider using a different model. If the null hypotheses in the type 2 or 3 plots are violated, consider using bootstrap p-values; see Section 8.1.5 of Thulin (2024) for details and recommendations.
References
Buja, Cook, Hofmann, Lawrence, Lee, Swayne, Wickham. (2009). Statistical inference for exploratory data analysis and model diagnostics, Phil. Trans. R. Soc. A, 367, 4361-4383.
Thulin, M. (2024) Modern Statistics with R. Boca Raton: CRC Press. ISBN 9781032512440. https://www.modernstatisticswithr.com/
See also
null_lm
Examples
data(tips)
x <- lm(tip ~ total_bill, data = tips)
lineup_residuals(x, type = 1) # Residuals vs Fitted
#> decrypt("gwqp 2J5J c0 t8Zc5c80 Ad")
 lineup_residuals(x, type = 2, method = "pboot") # Normal Q-Q plot
#> decrypt("gwqp 2J5J c0 t8Zc5c80 Aa")
lineup_residuals(x, type = 2, method = "pboot") # Normal Q-Q plot
#> decrypt("gwqp 2J5J c0 t8Zc5c80 Aa")
 lineup_residuals(x, type = 4) # Residuals vs Leverage
#> decrypt("gwqp 2J5J c0 t8Zc5c80 AA")
lineup_residuals(x, type = 4) # Residuals vs Leverage
#> decrypt("gwqp 2J5J c0 t8Zc5c80 AA")
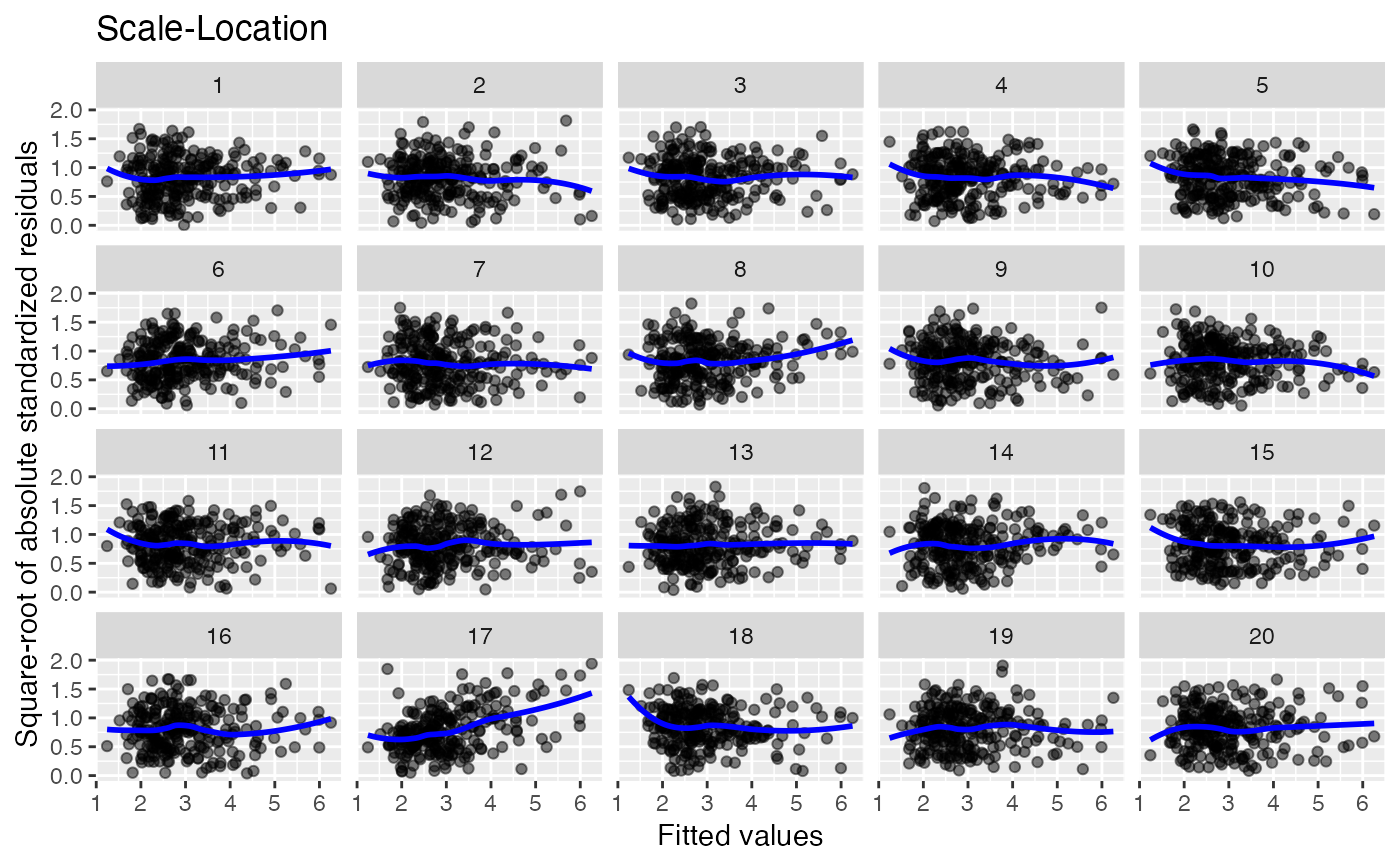 # Style the plot using color settings and ggplot2 functions:
lineup_residuals(x, type = 3,
color_points = "skyblue",
color_trends = "darkorange") +
ggplot2::theme_minimal()
#> decrypt("gwqp 2J5J c0 t8Zc5c80 da")
# Style the plot using color settings and ggplot2 functions:
lineup_residuals(x, type = 3,
color_points = "skyblue",
color_trends = "darkorange") +
ggplot2::theme_minimal()
#> decrypt("gwqp 2J5J c0 t8Zc5c80 da")